Abstract
Background
MYCN amplification is a prognostic biomarker associated with poor prognosis of neuroblastoma in children. The overall survival of children with MYCN-amplified neuroblastoma has only marginally improved within the last 20 years. The Bromodomain and Extra-Terminal motif (BET) inhibitor, JQ1, has been shown to downregulate MYCN in neuroblastoma cells.
Objective
To determine if JQ1 downregulation of MYCN in neuroblastomas can offer a target- specific therapy for this, difficult to treat, pediatric cancer.
Methods
Since MYCN-amplified neuroblastoma accounts for as much as 40 to 50 percent of all high-risk cases, we compared the effect of JQ1 on both MYCN-amplified and non-MYCN-amplified neuroblastoma cell lines and investigated its mechanism of action.
Results
In this study, we show that JQ1 can specifically target MYCN for downregulation, though this effect is not specific to only MYCN-amplified cells. And although we can confirm that the loss of MYCN alone can induce apoptosis, the exogenous rescue of MYCN expression can abrogate much of this cytotoxicity. More fascinating, however, was the discovery that the JQ1-induced knockdown of MYCN, which led to the loss of the human double minute 2 homolog (HDM2) protein, also led to the accumulation of tumor protein 53 (also known as TP53 or p53), which ultimately induced apoptosis. Likewise, the knockdown of p53 also blunted the cytotoxic effects of JQ1.
Conclusion
These data suggest a mechanism of action for JQ1 cytotoxicity in neuroblastomas and offer a possible prognostic target for determining its efficacy as a therapeutic.
Keywords: Cancer, oncology, neuroblastoma, BET inhibitors, p53, pediatric, MYCN amplifications, JQ1
1. INTRODUCTION
Neuroblastoma is a childhood cancer that commonly develops along the sympathetic nervous system or adrenal glands and affects approximately 1/7000 children [1]. It is the most commonly diagnosed malignancy in infants, accounting for 6% of all childhood cancers, but causing a disproportionally high 15% of all childhood cancer deaths in the United States [2-4]. While a small subset of neuroblastoma will undergo spontaneous regression, the long-term survival of patients with high-risk tumor phenotypes is less than 40% [5, 6].
The most common treatments for neuroblastoma include surgery and radiation therapy and/or chemotherapy [7, 8] with common complications including pulmonary problems, retardation of growth, vertebral deformity, renal impairment, second neoplasms, cardiac toxicity osteoporosis, and thyroid dysfunction, among others [9, 10]. And although new therapeutic techniques exist for patients with recurrent and refractory neuroblastoma (such as anti-angiogenesis therapy, immunotherapy, stem cell/bone marrow transplantation, etc.), many of these therapies have the disadvantages of toxic side effects, induction of early resistance, low specificity, and high cost [11-15].
Within the tumor subpopulation of neuroblastoma, a challenging heterogeneity exists, with prognostic factors for survival, including the age at diagnosis, both the tumor site and grade, histological analysis, and the amplification of the virus-Myelocytomatosis (or v-myc) oncogene neuroblastoma derived homolog gene, n-myc (also known as MYCN) [5, 6]. MYCN amplification is one of the most significant biomarkers, correlating with both advanced disease and poor survival, with as much as 20% - 25% of patients containing the MYCN amplification [16, 17].
Bromodomain and Extra-Terminal motif (BET) inhibitors are small molecules, which competitively displace BET bromodomain proteins from the chromatin by binding to acetyl-lysine recognition regions [18]. This BET protein binding inhibition leads to transcriptional target gene downregulation and has steered attention to these small molecules as putative cancer therapeutics [19, 20]. One particular BET inhibitor, JQ1, gained interest from its ability to inhibit Bromodomain-containing protein 3 (BRD3) and Bromodomain-containing protein 4 (BRD4), which form fusion oncogenes that drive NUT midline carcinoma [18, 21]. Since then, additional interest has arisen in other cancers that showed sensitivity to BET inhibitors, such as multiple myeloma, acute lymphoblastic leukemia, and acute myelogenous leukemia [22-24]. In addition, BET inhibitors have been explored as therapies for heart diseases, HIV infection, and even as a male contraceptive [25-27].
JQ1 is a thienotriazolodiazepine, a heterocyclic compound containing a diazepine ring fused to thiophene and triazole rings, and is structurally related to benzodiazepines (doi:10.1093/chromsci/ 36.3.111). Recent evidence has suggested that (+)-JQ1, but not (-)-JQ1 enantiomer, specifically targets MYCN in neuroblastomas [28]. Competitive binding of (+)-JQ1 to the MYCN promoter has shown to displace BRD4 from enhancer binding sites, leading to the downregulation of MYCN and inducing apoptotic cell death. Puissant et al. reported that MYCN-amplification in neuroblastomas was key to the reported cytotoxicity, however, a direct correlation between the knockdown of MYCN by JQ1 and apoptosis was never made [28]. Likewise, the mechanism of action of JQ1-induced apoptosis was never identified.
To that end, we decided to examine the activity of JQ1 in a panel of neuroblastomas. Our results indicate that MYCN-amplification is not necessary for the cytotoxic effects induced by JQ1, though the loss of MYCN expression is key to JQ1-induced apoptosis. Likewise, the exogenous rescue of MYCN expression can offset much of the cytotoxicity witnessed. In addition, JQ1 application confirmed that the loss of MYCN led to the loss of its transcriptional target, Human Double Minute 2 homolog (HDM2), an E3 ubiquitin ligase known to target tumor protein 53 (also known as TP53 or p53) [29, 30]. p53 is a tumor-suppressor protein whose functions are induced to activate cell cycle arrest or induce cell death through apoptosis [31, 32]. Loss of HDM2 crippled its ability to post-translationally regulate p53. This allowed for the accumulation of p53 and induction of apoptosis. Knockdown of p53 also rescued cellular cytotoxicity, offering a putative mechanism of action for JQ1-induced apoptosis in neuroblastoma cells. Together, our data indicates that JQ1 could be effective as a therapeutic agent against any neuroblastoma expressing MYCN, though p53 activity could be critical to the efficacy of this treatment.
2. METHODS
2.1. Cell Culture
LA-N-6, SK-N-Be (1), and SMS-KAN cells were cultured in HyClone RPMI-1640 [GE Healthcare Life Sciences] supplemented with 10% Fetal Bovine Serum (FBS). CHLA-42 cells were cultured in HyClone Iscove's Modified Dulbecco's Medium (IMDM) [GE Healthcare Life Sciences] supplemented with 20% FBS and 1X ITS (5μg/mL insulin, 5μg/mL transferrin, 5ng/mL selenium). IMR-32 cells were cultured in Minimum Essential Medium (MEM) Alpha + GlutaMAX™ [Gibco Life Sciences] supplemented with 10% Fetal Bovine Serum [FBS]. SK-N-AS cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) [Gibco Life Sciences] supplemented with 10% FBS and 1% Non-Essential Amino Acids (NEAA). CHLA-42, LA-N-6, SMS-KAN, and SK-N-Be (1) cells were all obtained from the Children's Oncology Group (Columbus, OH). Both IMR-32 and SK-N-AS cells were purchased from ATCC. All cells were incubated and maintained at 37oC with 5% CO2.
In order to confirm the identity of the cell lines obtained, they were screened for biological markers used to originally identify these neuroblastoma cells (Supplemental Table 1 (857.1KB, pdf) ) [33-38]. This was performed by examining the gene expression profiles of CDKN2A (Supplemental Fig. 1A (857.1KB, pdf) ) [39] and p73 (Supplemental Fig. 1B (857.1KB, pdf) ) [40], as well as MYCN [41], and p53 [42]. Validation of the proper phenotypes by this panel corroborated the viability of the cell lines used in this endeavor. The cell lines were thawed and passaged twice prior to validation and experimentation.
2.2. Cell Line Treatments with JQ1
All neuroblastoma cell lines were treated with either (+)-JQ1 or (-)-JQ1 enantiomer (molecular structures indicated in Supplemental Fig. 2A (857.1KB, pdf) , B).
Experimentally, 8x103 of each cell line were seeded in a volume of 100µL into 30 wells of a flat-bottom 96-well tissue culture treated plate and allowed to attach overnight (1 plate per cell line). The following day, cells were treated with either (+)-JQ1 or (-)-JQ1 enantiomer at concentrations of 0, 1, 2, 4, or 8µM in sextuplicate (6 wells per dose per cell line). All experiments were performed as triplicate independent measurements. All cells were then maintained for 72 hours at 37oC with 5% CO2.
2.3. Cell Viability Assay
Using the cells plated and treated with JQ1 after 72 hours (as above in “Cell line treatments with JQ1”), a CellTiter 96® Aqueous One Solution Cell Proliferation (Promega Corp.; Madison, WI) assay was performed, according to the manufacturer’s instructions (PerkinElmer Multilabel Plate Reader –Model 2104) with each well measured in quadruplicate. The results were determined from experiments performed in triplicate from triplicate independent measurements.
2.4. Bright Field Examination of Neuroblastoma Cells Following JQ1 Treatment
Each neuroblastoma cell line was washed with 1x Phosphate Buffered Saline (PBS), then lifted and dissociated using Accumax solution (Sigma-Aldrich). The cells were counted using a Cellometer (Nexcelom Bioscience) and 2.8x105 cells of each neuroblastoma cell line were plated into two wells of a 12-well tissue culture plate. Each sample was either treated with 0 (no treatment) or 8µM JQ1, then incubated at 37°C in 5% CO2. After 48 hours, the plates were examined under bright field conditions using an EVOS M5000 imaging system (ThermoFisher Scientific). Images were examined at a magnification of 40x. The experiment was performed with triplicate independent treatments. Three representative images were acquired from each treatment from which a final representative image was selected.
2.5. Apoptosis Assay (Caspase 3/7)
Using the cells plated and treated with JQ1 after 72 hours (as above in “Cell line treatments with JQ1”), 100uL of Caspase-Glo® 3/7 (Promega Corp.) reagent was added to each well and allowed to incubate at room temperature for 2 hours. Caspase activity was measured for luminescence using a GloMax luminometer (Promega) with each well measured in quadruplicate, comparing JQ1-treated cells to JQ1-untreated cells. The results were determined from experiments performed in triplicate from triplicate independent measurements.
2.6. Quantitative Reverse Transcription-Polymerase Chain Reaction of Neuroblastoma Cell Lines
Total cellular RNA was isolated from cell samples using an RNeasy Mini Kit (Qiagen) and purified following the manufacturer’s instructions. In brief, cells were pelleted by centrifugation, washed with 1x PBS, then resuspended in QIAzol Lysis Reagent to homogenize the sample. Chloroform was added to each sample and mixed vigorously, then centrifuged at 12000xg for 15 minutes. The aqueous phase was then mixed with 100% ethanol and centrifuged through an RNeasy isolation column. The column was washed and the sample was eluted in RNase-free water. The RNA concentrations of each sample were determined by using a Nanodrop (Nanodrop 2000c spectrophotometer, ThermoFisher Scientific). mRNA was then converted into cDNA using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Real-Time quantitative Polymerase Chain Reaction (qRT-PCR) was performed using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories), using Power SYBR Green PCR Master Mix (Applied Biosystems, Thermo Scientific) to amplify samples in triplicate Gene expression values were determined from three independent measurements. Gene-specific qPCR primer sequences were as follows: GAPDH, sense primer, 5’-ACATCGCTCAGACACCATG-3’, and anti-sense primer, 5’-TGTAGTTGAGGTCAATGAAGGG-3’; MYCN, sense primer, 5’-GACCACAAGGCCCTCAGTACCTCC-3’, and anti-sense primer, 5’-CACAGTGACCACGTCGATTTCTTCC-3’; and TP53, sense primer, 5’-CTCAAGGATGCCCAGGCTGGG-3’, and anti-sense primer, 5’-TATGGCGGGAGGTAGACTGACCC-3’. The results were reported as means ±SEM.
2.7. Construction of MYCN Recombinant Expression Vector
Total RNA was isolated from IMR-32 cells using an RNeasy Mini Kit (Qiagen), as described in the above section “Quantitative Reverse Transcription-Polymerase Chain Reaction of Neuroblastoma cell lines”. Purified RNA was then reverse-transcribed using M-MLV reverse transcriptase (ThermoFisher Scientific, Cat# 4368814). The resulting cDNA was then used as a template for PCR amplification using GoTaq (Promega). The PCR product was gel purified using a QIAquick Gel Extraction kit (Qiagen) as follows: the PCR sample was loaded into the well of a 1% agarose gel and run for 30 minutes at 100v, using an All-Purpose Hi-Lo DNA Marker (Bionexus). The PCR product was visualized under UV light, cut from the gel, melted in a solubilization buffer, and centrifuged through a QIAquick Gel Extraction column. The column was then washed and the sample was eluted in 10mM Tris, pH 8.0. The eluate PCR product was TOPO-cloned into pCR4-TOPO (Life Technologies), transformed into Top10 Chem comp cells, and then plated onto LB Amp plates (100ug/mL). Colonies were grown in LB Amp (100ug/mL) overnight at 37oC. Plasmids were harvested by miniprep using QIAprep Spin Miniprep kit (Qiagen) as follows: bacterial cells were pelleted from cultured media by centrifugation, resuspended in P1 Resuspension buffer, lysed in P2 Lysis buffer, and neutralized in N3 buffer. The neutralized lysate was then centrifuged at 13000xg for 3 minutes and the aqueous lysate was centrifuged through a QIAprep spin column. The column was washed and the sample eluted in 10mM Tris, pH 8.0. Each isolated plasmid was submitted for sequencing (Retrogen) and then analyzed using VectorNTi and AlignX (Life Technologies). The MYCN clone was then sub-cloned into pcDNA6/V5-HisA by restriction digestion and ligated using T4 Ligase (NEB, Inc.). The insertion of MYCN into the final clone was confirmed by restriction digestion.
2.8. Stable Selection of Expression Vectors into IMR-32 Cells
IMR-32 cells were seeded into single wells of a 6-well plate at a density of 2.5x105 cells per well and transfected with either pcDNA6/V5-HisA (Vector Control) or pcDNA6/MYCN. In brief, FuGENE® 6 (Promega) was mixed with OPTI-MEM reduced serum media (ThermoFisher) and allowed to mix for 5 minutes at RT. 1ug of plasmid was then added to this solution, raising the total volume to 100uL, and allowed to mix for 15 minutes at RT. The transfection solution was then added dropwise to IMR-32 cells. The transfection was allowed to continue for 6 hours, after which the media was removed, the cells were washed with PBS, and fresh media was added. 24 hours after transfection, the cells were selected with 4ug/mL Blasticidin (Life Technologies Corp.). Selection continued for 10 days until individual colonies could be isolated.
2.9. Confirmation of Stable Expression of MYCN in IMR-32 Cells
2.5x105 cells of each IMR-32 cell line (to include WT IMR-32, IMR-32 /VO, and IMR-32 /MYCN) were acquired and counted. Total cellular RNA was isolated from these cells using an RNeasy Mini Kit (Qiagen), as described in the above section “Quantitative Reverse Transcription-Polymerase Chain Reaction of Neuroblastoma cell lines”. mRNA was then converted into cDNA (as the previous protocol) and Real-time quantitative PCR was performed as noted previously. Gene expression values were determined from three independent measurements. The results were reported as means ±SEM.
2.10. Analysis of Cellular Cytotoxicity
8x103 cells were plated in a flat-bottom 96-well tissue culture plate and treated with the CellTox™ Green Dye (Promega Corp.) reagent. The cells were then allowed to attach overnight. The following day, each cell line was treated with JQ1 at a concentration of 0 or 8µM in sextuplicate (6 wells per dose per cell line). The plates were incubated at room temperature for 15 minutes, shielded from ambient light, then shaken for 1 minute on an orbital shaker (700-900g). Fluorescence was then measured at 485-500nmEx/520-530nmEm. All cells were maintained at 37oC in 5% CO2. Every 24 hours, for 5 days, the plates were removed from the incubator, shaken, then measured for fluorescence according to the manufacturer’s instructions. Data presented are a composite of experiments performed in three independent measurements.
2.11. siRNA-Mediated Knock-Down in Neuroblastoma Cells
Neuroblastoma cells were seeded into single wells of a 6-well plate at a density of 2.5x105 cells per well and transfected with 50uM of gene-specific siRNA or Silencer Select Negative Control siRNA #1 (Cat # 4390843). In brief, 9uL Lipofectamine RNAiMax (ThermoFisher) was mixed with 150uL OPTI-MEM reduced serum media (ThermoFisher). Separately, 6uL siRNA solution was mixed with 150uL OPTI-MEM reduced serum media. The two solutions were combined and allowed to mix for 5 minutes at RT. The transfection solution was then added dropwise to the neuroblastoma cells. The transfection was allowed to continue for 6 hours, after which the media was removed, the cells were washed with PBS, and fresh media was added. Samples were harvested for total RNA after 48 hours as follows: Each neuroblastoma cell line was washed with 1x Phosphate Buffered Saline (PBS), then lifted and dissociated using Accumax solution (Sigma-Aldrich). Total cellular RNA was isolated from these cells using an RNeasy Mini Kit (Qiagen), as described in the above section “Quantitative Reverse Transcription-Polymerase Chain Reaction of Neuroblastoma cell lines”. The gene-specific siRNAs are as follows: MYCN Silencer Select Pre-designed siRNA (Cat # 4392420, ID: s9133) and TP53 Silencer Select Pre-designed siRNA (Cat # 4390824, ID: s607).
2.12. Western Blot Analyses of Neuroblastoma Cells
Harvested cells (as above) were collected and counted, then boiled in sample buffer containing Sodium Dodecyl Sulphate (SDS) and βeta-Mercaptoethanol (β-ME) and proteins separated by electrophoresis on 4-20% gradient Tris-Glycine denaturing polyacrylamide gels (Cat# XP04200BOX, ThermoFisher Scientific). Proteins were transferred to nitrocellulose membranes (0.2um, BioRad, Cat# 1620112) and probed with the following primary antibodies: Anti-TP53 (Cell Signaling Technology, Inc.) at 1/200, anti-HDM2 (MDM2) (clone 3G9, EMD Millipore) at 1/500, and anti-GAPDH (Santa Cruz, FL-335) at 1/2000. Blots were probed with horseradish peroxidase-conjugated secondary antibodies (Invitrogen, Goat anti-Mouse, Cat# 62-6520, Goat anti-Rb, Cat# 65-6120) and visualized with ECL chemiluminescence (Pierce).
3. RESULTS
3.1. An Array of Neuroblastoma Cells is Sensitive to JQ1 Treatment
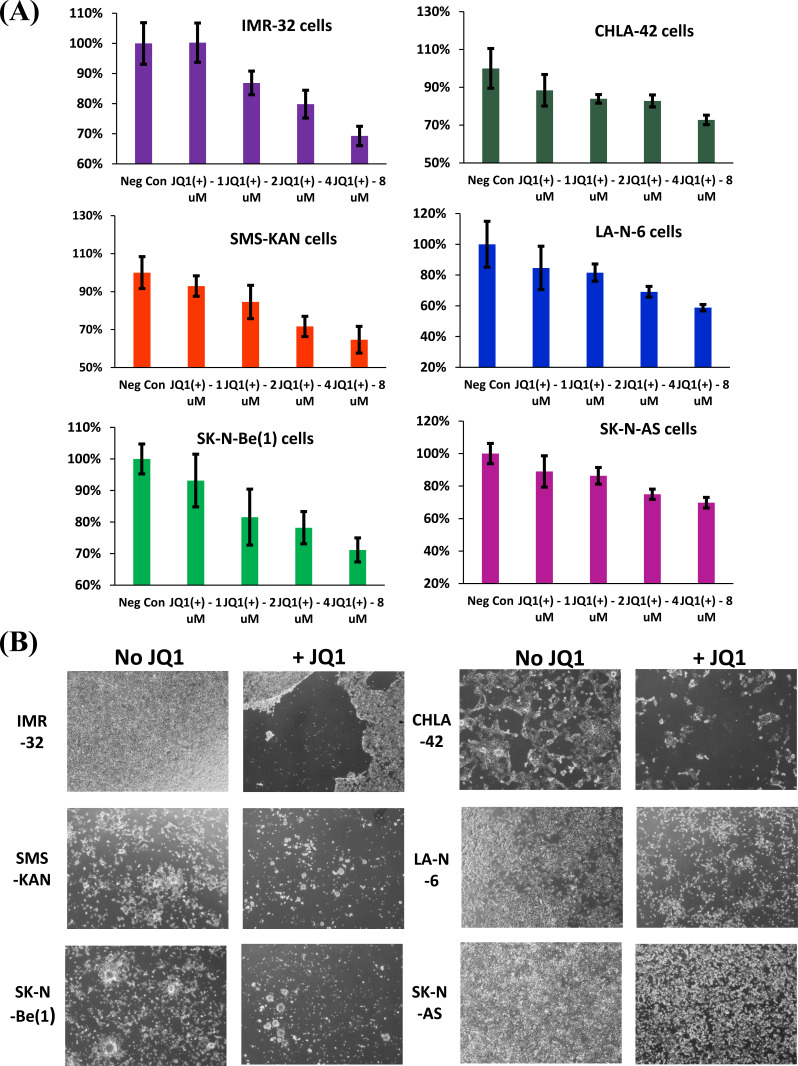
In order to determine the extent to which neuroblastoma cells are sensitive to JQ1, a panel of cultured neuroblastoma cells was treated, to include both MYCN-amplified cells [IMR-32, SMS-KAN, and SK-N-Be(1)] and non-MYCN-amplified cells (CHLA-42, LA-N-6, and SK-N-AS). The treatment was performed over a 48 hour period, using a gradient to include 0, 1, 2, 4, and 8uM concentrations of
both (+)-JQ1 and (-)-JQ1 enantiomer. All the samples were then measured for cell viability by MTS assay. The results indicated that all of the treated neuroblastoma cell lines exhibited sensitivity to (+)-JQ1 enantiomer, regardless of MYCN-amplification status (Fig. 1A). However, corroborating the results observed by Puissant et al. [28], SK-N-AS cells revealed the poorest response to JQ1, and bright field images confirmed only a marginal loss of cells compared to all the other neuroblastoma cell lines (Fig. 1B). It is worth noting that all of the cell lines screened for treatment by comparable concentrations of the (-)-JQ1 enantiomer revealed no significant loss in cell viability that could be discerned (data not shown). Regardless, these data suggest that SK-N-AS cells support a unique resistance to (+)-JQ1 treatment (hereafter referred to simply as “JQ1”).
Fig. (1).
Cell viability of human neuroblastoma cell lines after treatment with JQ1. (A) Three MYCN-amplified [IMR-32, SMS-KAN, and SK-N-Be(1)], and three non-MYCN amplified (CHLA-42, LAN-6, and SK-N-AS) neuroblastoma cell lines were treated with JQ1 at concentrations of 0 (no treatment), 1, 2, 4, or 8 µM for 48 hours. Cell viability (%) was assessed by MTS assay. Data shown are the composite of experiments performed in triplicate, with error bars representing standard deviation. (B) Bright-field images of the various neuroblastoma cell lines following treatment with either 0 (no treatment) or 8 µM JQ1 after 48 hours. Images were examined at a magnification of 40x. The experiment was performed with triplicate independent treatments. Three representative images were acquired from each treatment from which a final representative image was selected. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
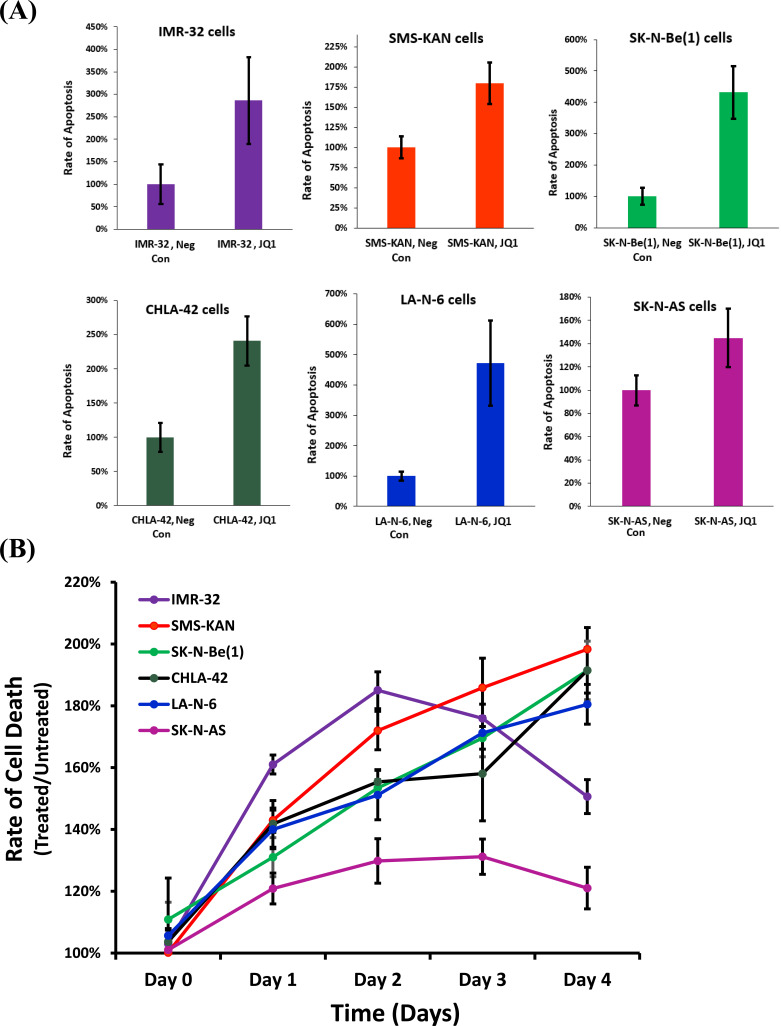
3.2. JQ1 Treatment Induces Apoptosis in all Tested Neuroblastoma Cell Lines, though Poorly in SK-N-AS Cells
Although all the neuroblastoma cell lines appeared sensitive to JQ1, the cellular viability of SK-N-AS cells was less effected. To further screen this phenotype, all the neuroblastoma cell lines were examined for changes in apoptosis after treatment with JQ1 for 48 hours, using the highest dose previously utilized (8uM) (Fig. 2A). Measurements in the changes of caspase 3/7 activity indicated dramatic increases in the rate of apoptosis, ranging from 180% of untreated (SMS-KAN) to as much as 500% of untreated (LA-N-6). Not surprisingly, SK-N-AS cells showed a significantly blunted effect on the rate of apoptosis after treatment, rising to only ~140% of untreated. In addition, measurements were taken utilizing a real-time cytotoxicity assay on the neuroblastoma cell lines after treatment with JQ1 (Fig. 2B). The results confirmed a suppressed effect of JQ1 on cellular cytotoxicity in SK-N-AS cells not witnessed in any of the other cell lines. These data indicate that SK-N-AS cells are resistant to the cytotoxic effects induced by JQ1 in neuroblastoma cells.
Fig. (2).
Apoptosis rate of human neuroblastoma cell lines after treatment with JQ1. Six neuroblastoma cell lines were evaluated comparing untreated vs. treated with JQ1 (8µM). (A) Apoptosis was assessed by measuring Caspase 3/7 activity. (B) Time Course of cellular cytotoxicity. Cell lines were treated with CellTox™ Green and JQ1 (8µM) with measurements taken every 24 hours for 4 days. The results were normalized to samples treated only with CellTox™ Green. Data for all experiments shown are the composite of experiments performed in triplicate from three independent measurements, with error bars representing standard deviation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.3. Sensitivity of Neuroblastoma Cells to JQ1 Directly Correlates with MYCN Expression and its Down-Regulation by JQ1, Though MYCN amplification is Not Necessary
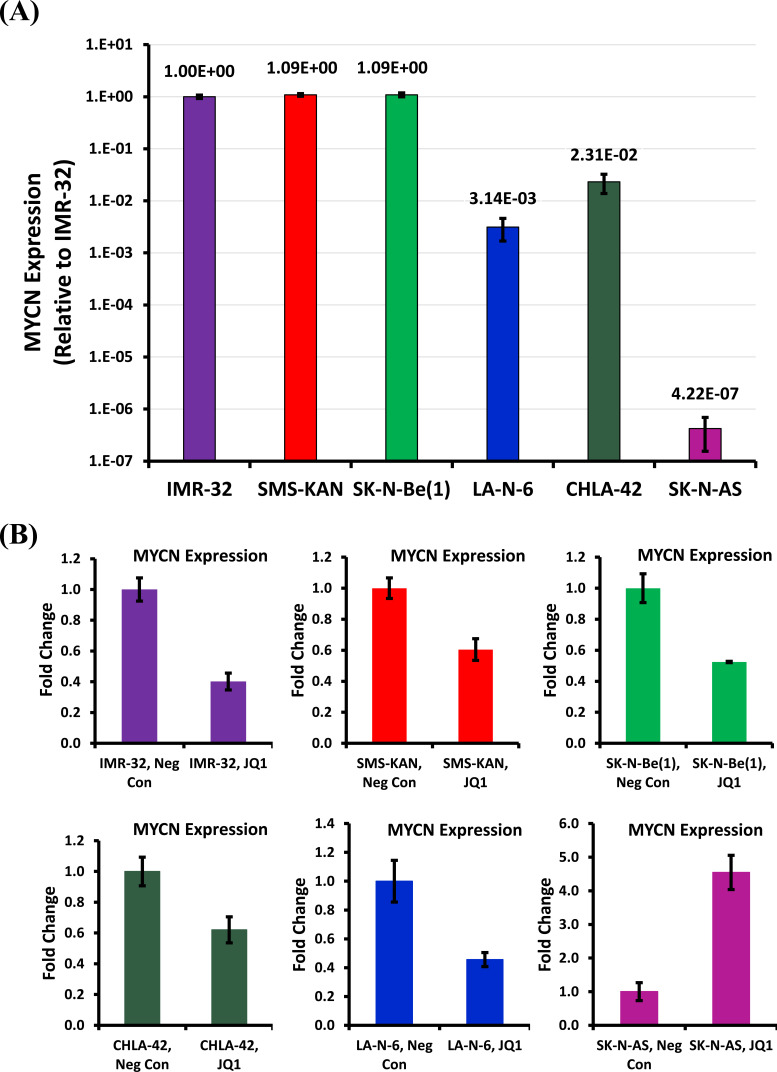
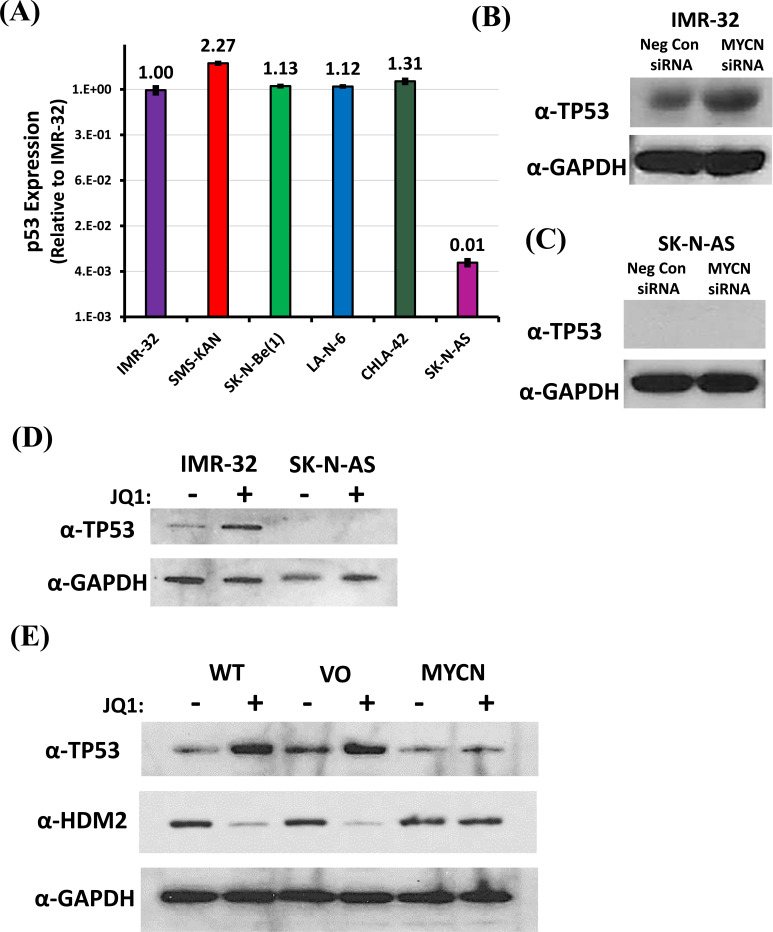
Given a dramatic resistance to JQ1 cytotoxicity in SK-N-AS cells, we decided to track down the possible cause. Puissant et al. concluded that JQ1 acted on neuroblastoma cells as a BET bromodomain inhibitor through the displacement of the BRD4 co-activator protein from both the BRD4 localization site as well as a putative enhancer region to the MYCN promoter [28]. Since this function of JQ1 explains its commensurate specificity to the MYCN promoter, it made sense to start by screening MYCN expression. An examination of MYCN expression in the neuroblastoma cell lines confirmed dramatically high expression of MYCN in the MYCN-amplified cell lines [IMR-32, SMS-KAN, and SK-N-Be(1)] (Fig. 3A). Both CHLA-42 and LA-N-6 also confirmed the expression of MYCN, though ~100-fold less than that seen in the MYCN-amplified cells, as expected. What was surprising, however, was the lack of MYCN expression present in the SK-N-AS cells, nearly 1000-fold less than that seen in the other non-MYCN-amplified cells, suggesting that it remains in a category of its own, that of MYCN down-regulated, not just non-amplified.
Fig. (3).
Analysis of MYCN expression in human neuroblastoma cell lines. (A) Relative MYCN mRNA expression was acquired from three MYCN-amplified [IMR-32, SMS-KAN, and SK-N-Be(1)] and three non-MYCN amplified (SK-N-AS, LAN-6, and CHLA-42) neuroblastoma cell lines by qRT-PCR. MYCN expression is normalized to GAPDH. (B) Analysis of MYCN expression was performed by qRT-PCR in the above neuroblastoma cell lines 48 hours after treatment, comparing untreated vs. JQ1 (8µM) treated cells. MYCN expression is normalized to GAPDH. All data shown are the composite of experiments performed in triplicate from three independent measurements, with error bars representing standard deviation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
It is interesting that measurements of MYCN expression after the treatment of JQ1 confirmed the down-regulation of MYCN in both the MYCN-amplified and non-MYCN-amplified cell lines (Fig. 3B). However, SK-N-AS expression of MYCN actually increased, suggesting that the MYCN response to JQ1 in these cells is also unique amongst the neuroblastoma cell lines. These data suggest that the effects of JQ1 on MYCN expression is consistent between both MYCN-amplified and non-MYCN-amplified cells, corroborating the phenotypes seen in cytotoxicity. However, the effects of JQ1 on MYCN expression are inconsistent in SK-N-AS cells, supporting the possibility that MYCN no longer influences cytotoxicity in a similar manner.
3.4. Exogenous Expression of MYCN can Rescue its Down-Regulation by JQ1 and Abrogate Cell Death Induced by JQ1 Treatment
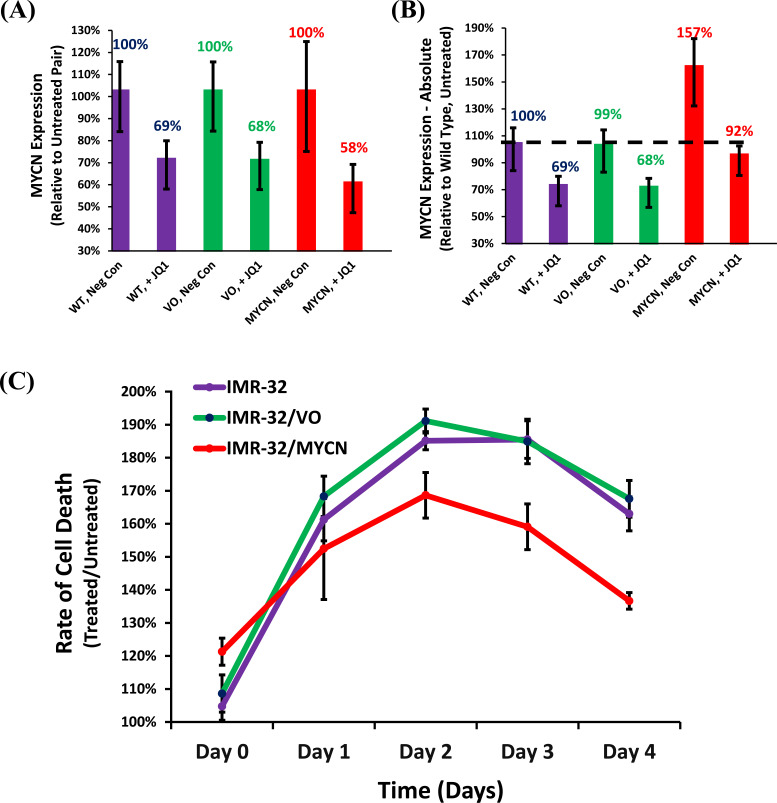
Since the cytotoxic responses to JQ1 in the majority of the neuroblastoma cell lines correlated with MYCN down-regulation, we sought to determine if this effect was direct or indirect. Therefore, a MYCN stably-expressing complimentary IMR-32 cell line was established. The MYCN expressed in this construct is driven by a promoter, which is not responsive to the BET bromodomain inhibition caused by JQ1. Treatment of these cells with JQ1 confirmed similar levels of MYCN knock-down to controls when measured proportional to its untreated pair (Fig. 4A). However, when measured relative to the untreated wild type, the levels of MYCN expression in the MYCN exogenously expressing cell line confirmed that it was present at ~160% of wild type when untreated, but at ~90% of wild type after treatment, suggesting that exogenous expression blunted the knock-down of MYCN caused by JQ1 treatment (Fig. 4B). A time course of real-time cytotoxicity confirmed that exogenous expression of MYCN could rescue a significant portion of the cytotoxicity caused by JQ1, confirming that the loss of MYCN expression alone could induce cell death in neuroblastoma cells (Fig. 4C). It is worth noting that exogenous expression of MYCN in IMR-32 cells did induce a basal increase in cytotoxicity of ~20% over controls, suggesting that the increase in MYCN expression was not harbored well in these cells. Thus, the advantage of increased MYCN expression was only witnessed when challenged by the knock-down of its expression.
Fig. (4).
Effect of JQ1 on neuroblastoma cell lines complimented with exogenous MYCN. (A) Comparison of Relative and (B) Absolute Quantification of MYCN expression by qRT-PCR. Cell lines examined include: IMR-32 Wild Type (WT), IMR-32 Vector only control (VO), and IMR-32 cells stably selected with exogenous expression of MYCN (MYCN). MYCN expression is normalized to GAPDH in all experiments. (C) Time Course of cellular cytotoxicity in the MYCN exogenously-expressing IMR-32 cells and their appropriate controls after treatment with JQ1. Cell lines were treated with CellTox™ Green and JQ1 (8µM), and cytotoxicity levels were measured every 24 hours for 4 days. The results were normalized to samples treated only with CellTox™ Green. Data shown are the composite of sextuplicate wells, performed in triplicate from three independent measurements, with error bars representing standard deviation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.5. JQ1 Down-Regulation of MYCN Leads to a Loss of HDM2, which Induces Stabilization of TP53 in Neuroblastoma Cells
In order to determine a possible mechanism to explain the resistance of SK-N-AS cells to JQ1 cytotoxicity, we compared them to the JQ1-sensitive IMR-32 cells. Previous work we performed [43] confirmed a discreet difference in both the mRNA and protein expression of Tumor Protein 53 (TP53 or p53) in IMR-32 cells when compared to SK-N-AS cells. A comparison of p53 amongst the neuroblastoma cell lines confirmed that its mRNA expression was comparable in all of the cell lines tested, except in SK-N-AS cells, which confirmed only ~1% of that seen in the other neuroblastoma cells (Fig. 5A). Treatment of IMR-32 cells with a MYCN-specific siRNA confirmed that specific knock-down of MYCN mRNA could induce an increase in the expression of p53 protein (Fig. 5B). However, p53 protein could not be detected in SK-N-AS cells, regardless of MYCN mRNA knock-down (Fig. 5C). Treatment of these cells with JQ1 confirmed similar results, with JQ1 inducing an increase in p53 protein levels in IMR-32 cells, but with no detectable levels of p53 protein in SK-N-AS cells regardless of the treatment (Fig. 5D).
Fig. (5).
Effect of MYCN downregulation on TP53 in neuroblastoma cells. (A) Relative TP53 mRNA expression was acquired from three MYCN-amplified [IMR-32, SMS-KAN, and SK-N-Be(1)] and three non-MYCN amplified (SK-N-AS, LAN-6, and CHLA-42) neuroblastoma cell lines by qRT-PCR. TP53 expression was normalized to GAPDH. Data shown are the composite of triplicate wells from three independent measurements. (B & C) Western blot analysis of the effect on TP53 protein in neuroblastoma cells (IMR-32 and SK-N-AS cells, respectively) treated with MYCN siRNA. Samples include Negative Control siRNA and MYCN siRNA. (D) Western blot analysis of the expression of TP53 protein in neuroblastoma cells (IMR-32 and SK-N-AS cells, respectively) treated with JQ1 (8 µM) compared to untreated cells. (E) Western blot analysis of the expression of TP53 and HDM2 (MDM2) proteins in Wild Type (WT), Vector only control (VO), and exogenously expressing MYCN (MYCN) IMR-32 cells treated with JQ1 (8µM) compared to untreated cells. Total protein analysis was performed compared to GAPDH as a control in all western blots. All data shown are the composite from three independent experiments. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Since MYCN exogenous expression was capable of rescuing the cytotoxicity of JQ1, p53 expression under those conditions was examined (Fig. 5E). The results confirmed that the rescue of MYCN expression also blunted the induction of p53. Although p53 is regulated transcriptionally, it is also abundantly regulated by post-translational ubiquitination, leading to its degradation [31, 44, 45]. Puissant et al. determined that the JQ1 knock-down of MYCN in neuroblastoma cells also led to the down-regulation of the MYCN target gene MDM2 (or HDM2, the human analog of this gene) [28]. Our previous results confirmed that HDM2 directly targets p53 in neuroblastoma cells, with the results leading to a direct loss of p53 protein [43]. An examination of HDM2 levels confirmed that JQ1 treatment of IMR-32 cells lead to a down-regulation of HDM2 (Fig. 5E). In addition, the exogenous rescue of
MYCN expression blunted the loss of HDM2. Together, these data suggest that the JQ1 treatment of neuroblastoma cells knocks down MYCN, leading to a loss of HDM2 expression, allowing for p53 protein accumulation.
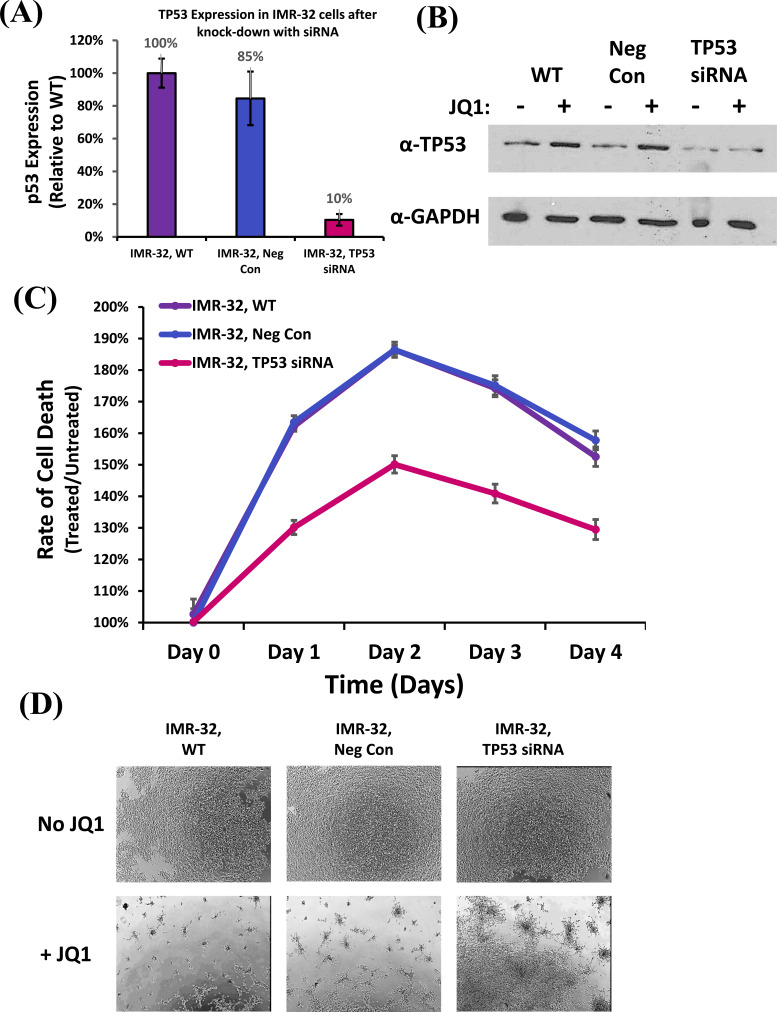
3.6. Knock-Down of TP53 can Rescue JQ1-Induced Cell Death in Neuroblastoma Cells
Although JQ1 treatment of neuroblastoma cells knocks down MYCN, which down-regulates HDM2, supporting the accumulation of p53, this does not directly confirm that p53 is responsible for inducing cell death. In order to determine whether p53 is directly responsible, we transfected IMR-32 cells with p53-specific siRNAs (Fig. 6A). The results confirmed by qRT-PCR that p53 mRNA was knocked-down by ~90% compared to controls. Likewise, western blot analysis confirms that cells transfected with p53-specific siRNAs produced less p53 protein than controls and could not induce a greater accumulation after treatment with JQ1 (Fig. 6B). These data support the hypothesis that the JQ1-dependent induction of p53 has been blunted.
Fig. (6).
Effect of TP53 on Cellular Cytotoxicity due to treatment with JQ1. (A) Relative TP53 mRNA expression was acquired from wild type IMR-32 cells (WT), compared to Negative Control siRNA-transfected (Neg Con), and TP53 siRNA-transfected (TP53 siRNA) cells, measured by qRT-PCR. TP53 expression was normalized to GAPDH. Data shown are the composite of triplicate wells from three independent measurements. (B) Western blot analysis of the expression of TP53 protein in wild type (WT), Negative Control siRNA-transfected (Neg Con), and TP53 siRNA-transfected samples (TP53 siRNA) IMR-32 cells treated with JQ1 (8µM) compared to untreated cells. Total protein analysis was performed compared to GAPDH as a control. (C) Time Course of cellular cytotoxicity in TP53-siRNA treated IMR-32 cell samples and their appropriate controls after treatment with JQ1. Cell lines were treated with CellTox™ Green and JQ1 (8 µM), and cytotoxicity levels were measured every 24 hours for 4 days. The results were normalized to samples treated only with CellTox™ Green. Data shown are the composite of sextuplicate wells, performed in triplicate from three independent measurements, with error bars representing standard deviation. (D) Brightfield images of TP53-siRNA treated IMR-32 cell samples and their appropriate controls following treatment with either 0 (no treatment) or 8 µM JQ1 after 48 hours. Images were examined at a magnification of 40x. Three representative images were acquired from each treatment from which a final representative image was selected. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
In order to experimentally establish whether the inhibition of p53 accumulation directly affects JQ1-dependent induction of cell death, we performed a real-time cytotoxicity. The results indicate that the knockdown of p53 in IMR-32 cells could rescue a significant portion of the cytotoxicity caused by JQ1 compared to control cells (Fig. 6C). Bright field images confirmed that p53-knock-down of IMR-32 cells improved cell survival, though some cell death still
occurred (Fig. 6D), suggesting that other mechanisms of action may still be at play.
CONCLUSION
While a small subset of neuroblastomas undergo spontaneous regression, about 70-80% of high-risk cases led to metastasis in as early as 18 months, with less than half experiencing remission [46]. Recently, Puissant et al. demonstrated the use of BET bromodomain inhibitors in high-risk MYCN-amplified neuroblastomas [28]. Their evidence indicated that MYCN is transcriptionally regulated by the BET protein BRD4, acting as a co-activator at putative enhancer regions in the MYCN promoter. Application of the bromodomain inhibitor JQ1 displaced BRD4 from its localization sites, repressing MYCN expression. Our studies confirmed that the JQ1 treatment of MYCN-amplified neuroblastoma cells resulted in the downregulation of MYCN as well as induction of apoptotic cell death, corroborating their data. However, the same results were also seen in non-MYCN-amplified cell lines, suggesting that MYCN-amplification is not required for this phenotype, merely moderate MYCN expression. And although we can confirm that SK-N-AS cells were significantly more resistant to JQ1 treatment, these cells do not express MYCN at levels comparable to other non-MYCN-amplified cells, but at levels many orders of magnitude below. In fact, JQ1 treatment of these cells actually increased MYCN expression, suggesting that not only phenotype but genotype is altered in this background. These results indicate that JQ1 could be considered for the treatment of a broader range of neuroblastomas beyond merely those that are MYCN-amplified. However, they also offer concerning implications for treatment, given that wild type MYCN expression in host neuroblasts could remain as likely a target as the neuroblastomas they intend to treat.
Although the correlation between BET bromodomain inhibition of MYCN expression by JQ1 was previously established, the mechanism of the downstream induction of apoptotic cell death in neuroblastomas was still left unclear. Previous work by Puissant et al. confirmed that the knockdown of MYCN by shRNA could induce apoptosis [28], however, this occurs through an alternate mechanism of action than JQ1. Rescue of MYCN expression resolved this concern, offering confirmation that the loss of MYCN expression alone in JQ1-treated neuroblastoma could induce majority of the cytotoxicity witnessed in these cells. Again, this corroborates the possibility of off-target effects caused by JQ1, given that any cell dependent upon MYCN expression could be adversely affected.
Of greater concern, however, is the mechanism of resistance to JQ1 witnessed in SK-N-AS cells. Previous work, we performed in neuroblastoma cells, identified the loss of TP53 in SK-N-AS cells, allowing them to resist induction of cell cycle arrest after the loss of the long non-coding RNA, GAS5 [43]. TP53 (or p53) is a well-known tumor suppressor often associated with induction of cell cycle arrest, though it also functions to induce cell death through apoptosis [31, 32, 44, 45]. A comparison of our panel of neuroblastoma cells confirmed that all the cell lines screened contained wild type levels of p53, except SK-N-AS, and knockdown of MYCN by siRNA or JQ1 treatment led to an increase in p53, except in SK-N-AS cells, where p53 could not be detected regardless of condition. The final key to this puzzle was actually presented by Puissant et al. [28], in their assessment of MYCN transcriptional targets affected by JQ1 treatment: the gene MDM2 (or HDM2 in humans). HDM2 is an E3 ubiquitin ligase known as a primary regulator of p53 [29, 30]. Treatment of neuroblastoma cells with JQ1 confirmed the loss of HDM2. And an exogenous expression of MYCN rescued not only the loss of HDM2, but inhibited the increase in p53. Likewise, a knockdown of p53 dramatically decreased the cytotoxic effects of JQ1, mirroring the phenotype witnessed by the rescue of MYCN.
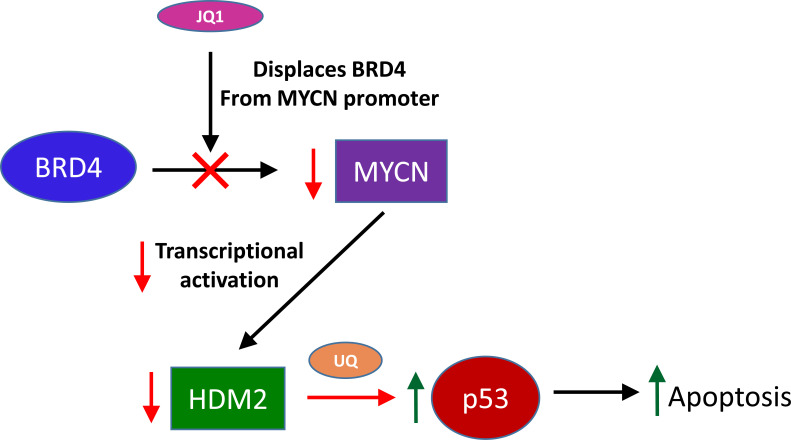
Together, these results allow us to establish a putative model for the network of action reflecting the phenotypes witnessed (Fig. 7). In short, the application of JQ1 displaces BRD4 from MYCN promoter enhancer sites, leading to decreased transcription of MYCN. The loss of MYCN leads to decreased transcription of HDM2, and to a loss of HDM2 protein. The loss of HDM2 protein diminishes its ability to regulate p53 protein, allowing for the accumulation of p53 in the cell, leading to the induction of apoptosis. Based upon this model, we suggest that, although MYCN may be a direct target of JQ1 inhibition, cytotoxicity is ultimately dependent upon p53 activity. Though it should be noted that both genes are critical to this network in neuroblastomas, corroborated by the results seen in SK-N-AS cells. Therefore, a critical prognostic of JQ1 efficacy in neuroblastomas might require the screening of p53 protein, in addition to MYCN amplification. Likewise, this quality might also explain the efficacy of JQ1 on other tumor types and the cause of some of its side effects as well.
Fig. (7).
Schematic illustration of the effects of JQ1 treatment on neuroblastoma cells. Briefly, JQ1 inhibits the binding of BRD4 proteins to enhancer sites in the MYCN promoter, leading to a reduction in the transcription of MYCN. This loss of MYCN expression leads to a decrease in the MYCN-dependent transcription of HDM2, leading to a loss of HDM2 protein. The loss of HDM2 protein reduces the ubiquitination of TP53, allowing TP53 protein to accumulate, inducing apoptosis in the host cell. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
The use of BET inhibitors, to treat human neuroblastomas, offers the advantage of selective targeting, through the downregulation of MYCN. Understanding that a portion of the cytotoxicity, induced through this process, occurs through p53-dependent apoptosis allows us to better understand its mechanism of action. In addition, we can now offer a possible prognostic marker to address the efficacy of JQ1 in neuroblastomas, allowing us to avoid unnecessary application and possibly improve the treatment outcome of this difficult to treat pediatric cancer.
ACKNOWLEDGEMENTS
We would like to thank the laboratory of Dr. Terri Finkel for their generous donation of both (+)-JQ1 and (-)-JQ1 enantiomers, without which none of these experiments could be performed. Additionally, we would like to thank both Jeanne Brooks and Amy Rosado for their assistance with qRT-PCR preparation and analysis.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
Funding for these studies was provided by the Nemours Foundation and Nemours Children’s Hospital (Grant No. 1014541001).
CONFLICT OF INTERESTS
The author declares no conflict of interest, financial or otherwise.
References
- 1.2017.
- 2.Brodeur G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 3.Maris J.M., Hogarty M.D., Bagatell R., Cohn S.L. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 4.Pandey G.K., Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. 2015;6(21):18265–18275. doi: 10.18632/oncotarget.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaioannou G., McHugh K. Neuroblastoma in childhood: Review and radiological findings. Cancer Imaging. 2005;5:116–127. doi: 10.1102/1470-7330.2005.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohn S.L., Pearson A.D., London W.B., Monclair T., Ambros P.F., Brodeur G.M., Faldum A., Hero B., Iehara T., Machin D., Mosseri V., Simon T., Garaventa A., Castel V., Matthay K.K. INRG Task Force. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis C.U., Shohet J.M. Neuroblastoma: Molecular pathogenesis and therapy. Annu. Rev. Med. 2015;66:49–63. doi: 10.1146/annurev-med-011514-023121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward E., DeSantis C., Robbins A., Kohler B., Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014;64(2):83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 9.Gurney J.G. Neuroblastoma, childhood cancer survivorship, and reducing the consequences of cure. Bone Marrow Transplant. 2007;40(8):721–722. doi: 10.1038/sj.bmt.1705815. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda T., Saeki M., Honna T., Kumagai M., Masaki H. Late complications after surgery in patients with neuroblastoma. J. Pediatr. Surg. 2006;41(12):2037–2040. doi: 10.1016/j.jpedsurg.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Tonini G.P., Pistoia V. Molecularly guided therapy of neuroblastoma: a review of different approaches. Curr. Pharm. Des. 2006;12(18):2303–2317. doi: 10.2174/138161206777585193. [DOI] [PubMed] [Google Scholar]
- 12.Alferiev I.S., Iyer R., Croucher J.L., Adamo R.F., Zhang K., Mangino J.L., Kolla V., Fishbein I., Brodeur G.M., Levy R.J., Chorny M. Nanoparticle-mediated delivery of a rapidly activatable prodrug of SN-38 for neuroblastoma therapy. Biomaterials. 2015;51:22–29. doi: 10.1016/j.biomaterials.2015.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alili L., Sack M., von Montfort C., Giri S., Das S., Carroll K.S., Zanger K., Seal S., Brenneisen P. Downregulation of tumor growth and invasion by redox-active nanoparticles. Antioxid. Redox Signal. 2013;19(8):765–778. doi: 10.1089/ars.2012.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari M., Singh S.P., Chinde S., Rahman M.F., Mahboob M., Grover P. Toxicity study of cerium oxide nanoparticles in human neuroblastoma cells. Int. J. Toxicol. 2014;33(2):86–97. doi: 10.1177/1091581814522305. [DOI] [PubMed] [Google Scholar]
- 15.Lin W., Huang Y.W., Zhou X.D., Ma Y. Toxicity of cerium oxide nanoparticles in human lung cancer cells. Int. J. Toxicol. 2006;25(6):451–457. doi: 10.1080/10915810600959543. [DOI] [PubMed] [Google Scholar]
- 16.Huang M., Weiss W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013;3(10):a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irwin M.S., Park J.R. Neuroblastoma: Paradigm for precision medicine. Pediatr. Clin. North Am. 2015;62(1):225–256. doi: 10.1016/j.pcl.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., Philpott M., Munro S., McKeown M.R., Wang Y., Christie A.L., West N., Cameron M.J., Schwartz B., Heightman T.D., La Thangue N., French C.A., Wiest O., Kung A.L., Knapp S., Bradner J.E. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., Chesi M., Schinzel A.C., McKeown M.R., Heffernan T.P., Vakoc C.R., Bergsagel P.L., Ghobrial I.M., Richardson P.G., Young R.A., Hahn W.C., Anderson K.C., Kung A.L., Bradner J.E., Mitsiades C.S. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott C.J., Kopp N., Bird L., Paranal R.M., Qi J., Bowman T., Rodig S.J., Kung A.L., Bradner J.E., Weinstock D.M. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz B.E., Hofer M.D., Lemieux M.E., Bauer D.E., Cameron M.J., West N.H., Agoston E.S., Reynoird N., Khochbin S., Ince T.A., Christie A., Janeway K.A., Vargas S.O., Perez-Atayde A.R., Aster J.C., Sallan S.E., Kung A.L., Bradner J.E., French C.A. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011;71(7):2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belkina A.C., Denis G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 2012;12(7):465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J., Vakoc C.R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 2014;54(5):728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Da Costa D., Agathanggelou A., Perry T., Weston V., Petermann E., Zlatanou A., Oldreive C., Wei W., Stewart G., Longman J., Smith E., Kearns P., Knapp S., Stankovic T. BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia. Blood Cancer J. 2013;3:e126. doi: 10.1038/bcj.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand P., Brown J.D., Lin C.Y., Qi J., Zhang R., Artero P.C., Alaiti M.A., Bullard J., Alazem K., Margulies K.B., Cappola T.P., Lemieux M., Plutzky J., Bradner J.E., Haldar S.M. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee C., Archin N., Michaels D., Belkina A.C., Denis G.V., Bradner J., Sebastiani P., Margolis D.M., Montano M. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J. Leukoc. Biol. 2012;92(6):1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzuk M.M., McKeown M.R., Filippakopoulos P., Li Q., Ma L., Agno J.E., Lemieux M.E., Picaud S., Yu R.N., Qi J., Knapp S., Bradner J.E. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puissant A., Frumm S.M., Alexe G., Bassil C.F., Qi J., Chanthery Y.H., Nekritz E.A., Zeid R., Gustafson W.C., Greninger P., Garnett M.J., McDermott U., Benes C.H., Kung A.L., Weiss W.A., Bradner J.E., Stegmaier K. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3(3):308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10(4):400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 30.Jain A.K., Barton M.C. Making sense of ubiquitin ligases that regulate p53. Cancer Biol. Ther. 2010;10(7):665–672. doi: 10.4161/cbt.10.7.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L., Tweddle D.A. p53, SKP2, and DKK3 as MYCN target genes and their potential therapeutic significance. Front. Oncol. 2012;2:173. doi: 10.3389/fonc.2012.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okoshi R., Ando K., Suenaga Y., Sang M., Kubo N., Kizaki H., Nakagawara A., Ozaki T. Transcriptional regulation of tumor suppressor p53 by cAMP-responsive element-binding protein/AMP-activated protein kinase complex in response to glucose deprivation. Genes Cells. 2009;14(12):1429–1440. doi: 10.1111/j.1365-2443.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 33.Tumilowicz J.J., Nichols W.W., Cholon J.J., Greene A.E. Definition of a continuous human cell line derived from neuroblastoma. Cancer Res. 1970;30(8):2110–2118. [PubMed] [Google Scholar]
- 34.El-Badry O.M., Romanus J.A., Helman L.J., Cooper M.J., Rechler M.M., Israel M.A. Autonomous growth of a human neuroblastoma cell line is mediated by insulin-like growth factor II. J. Clin. Invest. 1989;84(3):829–839. doi: 10.1172/JCI114243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada R.K., Seeger R.C., Brodeur G.M., Einhorn P.A., Rayner S.A., Tomayko M.M., Reynolds C.P. Human neuroblastoma cell lines that express N-myc without gene amplification. Cancer. 1993;72(11):3346–3354. doi: 10.1002/1097-0142(19931201)72:11<3346:AID-CNCR2820721134>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.Keshelava N., Seeger R.C., Groshen S., Reynolds C.P. Drug resistance patterns of human neuroblastoma cell lines derived from patients at different phases of therapy. Cancer Res. 1998;58(23):5396–5405. [PubMed] [Google Scholar]
- 37.Reynolds C.P., Biedler J.L., Spengler B.A., Reynolds D.A., Ross R.A., Frenkel E.P., Smith R.G. Characterization of human neuroblastoma cell lines established before and after therapy. J. Natl. Cancer Inst. 1986;76(3):375–387. [PubMed] [Google Scholar]
- 38.Biedler J.L., Roffler-Tarlov S., Schachner M., Freedman L.S. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. 1978. [PubMed]
- 39.Thompson P.M., Maris J.M., Hogarty M.D., Seeger R.C., Reynolds C.P., Brodeur G.M., White P.S. Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res. 2001;61(2):679–686. [PubMed] [Google Scholar]
- 40.Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J.C., Valent A., Minty A., Chalon P., Lelias J.M., Dumont X., Ferrara P., McKeon F., Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–819. doi: 10.1016/S0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 41.Ciccarone V., Spengler B.A., Meyers M.B., Biedler J.L., Ross R.A. Phenotypic diversification in human neuroblastoma cells: Expression of distinct neural crest lineages. Cancer Res. 1989;49(1):219–225. [PubMed] [Google Scholar]
- 42.Keshelava N., Zuo J.J., Chen P., Waidyaratne S.N., Luna M.C., Gomer C.J., Triche T.J., Reynolds C.P. Loss of p53 function confers high-level multidrug resistance in neuroblastoma cell lines. Cancer Res. 2001;61(16):6185–6193. [PubMed] [Google Scholar]
- 43.Mazar J., Rosado A., Shelley J., Marchica J., Westmoreland T.J. The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget. 2017;8(4):6589–6607. doi: 10.18632/oncotarget.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May P., May E. Twenty years of p53 research: Structural and functional aspects of the p53 protein. Oncogene. 1999;18(53):7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 45.Ko L.J., Prives C. p53: Puzzle and paradigm. Genes Dev. 1996;10(9):1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 46.Look A.T., Hayes F.A., Shuster J.J., Douglass E.C., Castleberry R.P., Bowman L.C., Smith E.I., Brodeur G.M. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: A Pediatric Oncology Group study. J. Clin. Oncol. 1991;9(4):581–591. doi: 10.1200/JCO.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
Not applicable.