Fig. 4.
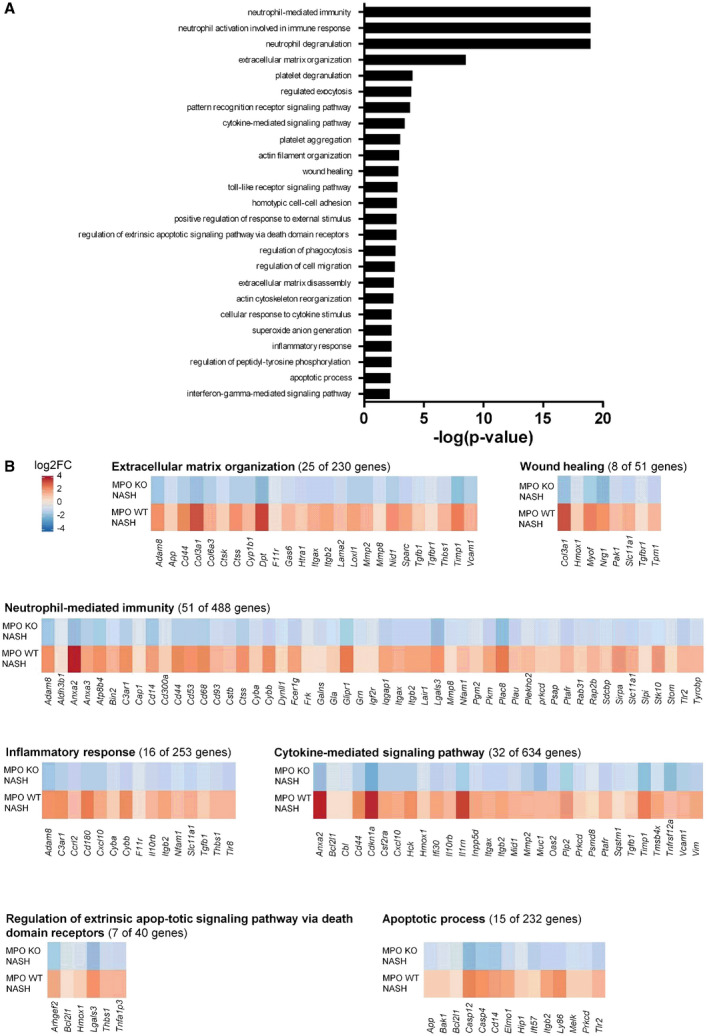
Transcriptome analysis of MPO‐dependent pathways in NASH. Microarray analysis from whole‐liver tissue of hepatic genes differentially (P < 0.05) expressed in WT mice compared with MPO KO animals following 24 weeks of HFCholC feeding. (A) Displayed are the top 25 MPO‐regulated pathways in NASH. (B) Gene‐expression heat maps for selected pathways. The heat maps display genes that were significantly (adjusted P value < 0.05) up‐regulated in the livers of WT mice with HFCholC‐induced NASH compared with chow‐fed WT mice (bottom panel) and down‐regulated in livers of MPO KO mice with HFCholC‐induced NASH compared with HFCholC‐fed WT mice (upper panel). Abbreviations: Adam 8; a disintegrin and metalloproteinase domain‐containing protein 8; Aldh3b1, aldehyde dehydrogenase 3 family, member B1; Anxa, annexin; App, amyloid precursor protein; Atp8b4, ATPase phospholipid transporting 8B4; Bcl2l1, Bcl‐2‐like protein 1; C3ar1, complement component 3a receptor; Cap1, cyclase associated actin cytoskeleton regulatory protein 1; Cbl, Cbl proto‐oncogene; Ccrl2, C‐C chemokine receptor‐like 2; Cd, cluster of differentiation; Cdkn1a, cyclin‐dependent kinase inhibitor 1; Col3a1, α1 type III collagen; Col6a1, α1 type VI collagen, Csf2ra, granulocyte‐macrophage colony‐stimulating factor receptor; Cstb, cystatin b; Ctsk, cathepsin K; Ctss, cathepsin S; Cxcl10, C‐X‐C motif chemokine 10; Cyba; cytochrome b‐245 α chain; Cybb, cytochrome b‐245 β chain; Cyp1b1, cytochrome P450 family 1 subfamily B member 1; Dpt, dermatopontin; Dynll1, dynein light chain LC8‐type 1; F11r, junctional adhesion molecule A; Fcer1g, Fc fragment of IgE, high affinity I, receptor for, gamma polypeptide; Frk, fyn‐related kinase; Galns, N‐acetylgalactosamine‐6‐sulfatase; Gas6, growth arrest specific 6; Gla, galactosidase α; Glipr1, glioma pathogenesis‐related protein 1; Grn, granulin; Hck, tyrosine‐protein kinase HCK; Hmox1, heme oxygenase 1; Htra1, HtrA serine peptidase 1; Ifi30, IFI30 lysosomal thiol reductase; Igf2r, insulin‐like growth factor 2 receptor; Il10rb, interleukin 10 receptor β subunit; Il1rn, interleukin 1 receptor antagonist; Inpp5d, inositol polyphosphate‐5‐phosphatase D; Iqgap1, IQ motif containing GTPase activating protein 1; Itgax, integrin subunit αX; Itgb1, integrin subunit β2; Lair1, leukocyte‐associated immunoglobulin‐like receptor 1; Lama2, laminin subunit α2; Lgals3, galectin 3; Loxl1, lysyl oxidase‐like 1; Mid1, midline 1; Mmp, matrix metalloproteinase; Muc1, mucin 1, cell surface associated; Myof, myoferlin; Nfam1, NFAT activating protein with ITAM motif 1; Nid1, nidogen 1; Nrg1, neuregulin 1; Oas2, 2'‐5'‐oligoadenylate synthetase 2; Pak1, p21‐activated kinase 1; Pgm2, phosphoglucomutase 2; Pkm, pyruvate kinase M1/2; Plac8, placenta‐associated 8; Plau, Urokinase; Plekho2, pleckstrin homology domain containing O2; Plp2, proteolipid protein 2; Prkcd, protein kinase C δ; Psap, prosaposin; Psmd8, proteasome 26S subunit, non‐ATPase 8; Ptafr, platelet‐activating factor receptor; Rab31, Ras‐related protein Rab 31; Rap2b, Ras‐related protein Rap 2b; Sdcbp, syndecan binding protein; Slc11a1, solute carrier family 11 member 1; Slpi, secretory leukocyte peptidase inhibitor; Sparc, osteonectin; Sqstm1, sequestosome 1; Stk10, serine/threonine‐protein kinase 10; Stom, stomatin; Tgfbr, transforming growth factor β receptor; Thbs1, thrombospondin 1; Tlr, toll‐like receptor; Tmsb4x, thymosin β4; Tnfrsf12a, tumor necrosis factor receptor superfamily member 12A; Tpm1, Tropomyosin α1 chain; Tyrobp, transmembrane immune signaling adaptor TYROBP; Vcam1, vascular cell adhesion molecule 1; Vim, vimentin.
