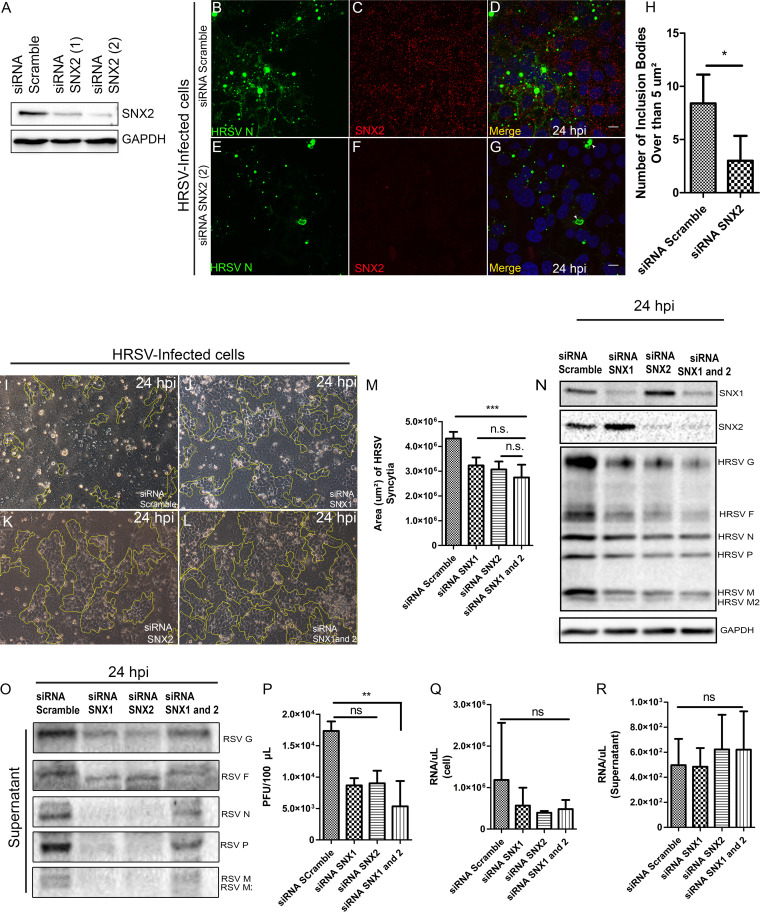
FIG 12.
Small interfering RNA for SNX1 and 2. (A) Immunoblotting for SNX2 in cells treated with siRNA for SNX2 after two shots (1 and 2) done with the same amount of siRNA; GAPDH was the load control. (B to D) Panel D shows a merge of panels B and C in cells treated with siRNA scrambled. (E to G: Panel G shows a merge of panels E and F in cells silenced for SNX2; arrowheads point to irregular-shaped inclusion bodies. (H) Graph of the inclusion body quantity over than 5 μm2 found in cell siRNA scramble and siRNA for SNX2 24 hpi. (I) Monolayer of HEp-2 cells, infected by HRSV and treated with scrambled siRNA. (J, K, and L) HRSV-infected HEp-2 monolayers treated with siRNA for SNX1, SNX2, and SNX1 and 2, respectively. The markings in the figures outline cell islands without syncytium formation. (M) Graph showing the sizes of HRSV-induced syncytia in cells treated with scrambled siRNA and siRNAs for SNX1, SNX2, and SNX1 and 2 at 24 hpi. (N) Immunoblotting for SNX1, SNX2, or both and for HRSV proteins, using HRSV-infected HEp-2 cells treated with scrambled siRNA or siRNA for SNX1, SNX2, or SNX1 and 2 at 24 hours postinfection. (O) Immunoblotting for HRSV proteins, using supernatants of HRSV-infected HEp-2 cells treated with scrambled siRNA or siRNA for SNX1, SNX2, or SNX1 and 2, at 24 hours postinfection. (P) HRSV progeny production in infected cells treated with scrambled siRNA or siRNA for SNX1, SNX2, or SNX1 and 2. (Q and R) HRSV RNA genome copies at 24 hours postinfection in cells and supernatants, respectively, in cultures treated with scrambled siRNA or with siRNA for SNX1, SNX2, or SNX1 and 2. The images are representative of a single focal plan, taken with a Zeiss 780 microscope. Magnification, ×63. This set of figures is representative of three independent experiments. The statistic of panel H was done using Student’s t test, and the graphs in panels M, P, Q, and R were done using ANOVA one way Tukey’s multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.