Figure 2.
Functional Analysis of Genes That Are Enriched or Depleted in Nuclei
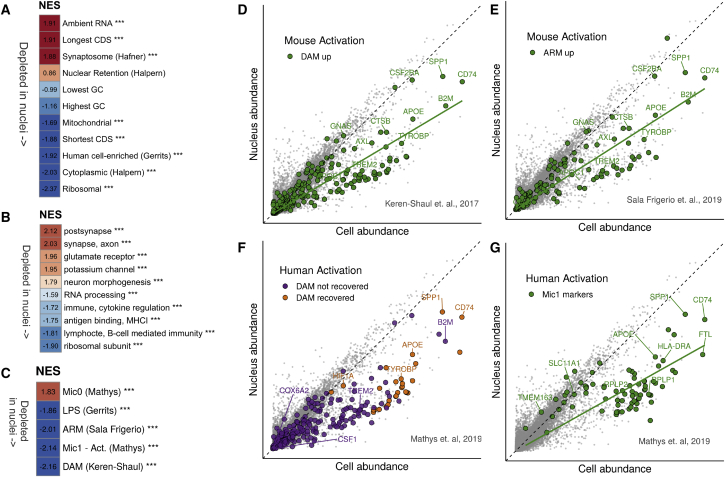
(A) Gene set enrichment analysis (GSEA) of gene sets related to cellular location and gene coding sequence (CDS) length. Background genes were ranked according to log fold change of nuclei (3,721 nuclei) versus cells (14,435 cells). Red, higher normalized enrichment score (NES), i.e., more genes associated with nuclear enrichment; blue, negative NES scores (depletion in nuclei). ∗∗∗ represents significance (padj < 0.0005). GC, GC content.
(B) GSEA of super-Gene Ontology gene sets against ranked nucleus-cell log fold changes. Only top and bottom categories (according to NES) are shown. Colors as in (A). MHCI, major histocompatibility complex class I.
(C) GSEA of selected gene sets from previous studies of microglial activation, against log fold change as in (A). ∗∗∗ represents significance (padj < 0.0005). Mic0, markers of microglial cluster 0 in human brain tissue; Mic1, markers of microglial cluster 1 (activation response to plaques) defined by Mathys et al., 2019 in human brain tissue. ARM, activation response microglia (Sala Frigerio et al., 2019); DAM, disease-associated microglia (Keren-Shaul et al., 2017); LPS, lipopolysaccharide (Gerrits et al., 2019).
(D) Scatterplot as in Figure 1A, highlighting in green the DAM genes. A regression line for the highlighted genes is shown in green (slope = 0.60).
(E) Scatterplot as in (D), highlighting in green the ARM genes. A regression line for the highlighted genes is shown in green (slope = 0.64).
(F) Scatterplot as in (D), highlighting the DAM genes recovered in the study of human activation in AD (Mathys et al., 2019). Purple, DAM genes not recovered in their study; orange, DAM genes recovered in their study.
(G) Scatterplot as in (D); green, human activation marker genes defined by Mathys et al. (2019). Gene sets, results of GO clustering, and results of GSEA analysis are available in Table S1. See also Figures S2B–S2G.