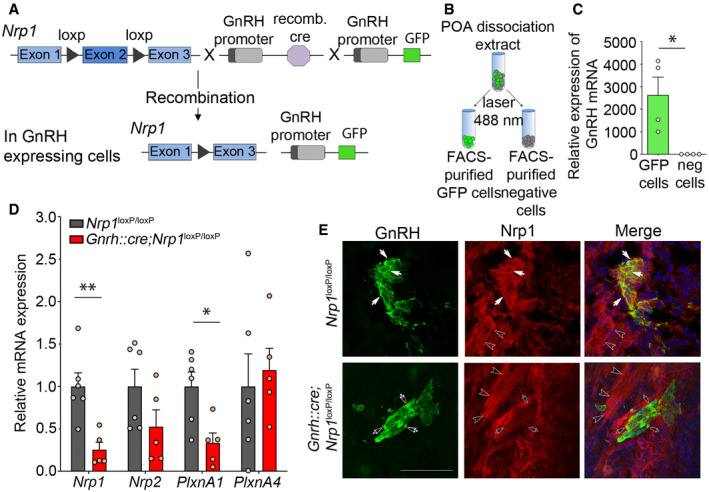
Genetic strategy to selectively knock out Nrp1 in GnRH‐expressing cells, using Nrp1
loxP/loxP mice crossed with Gnrh::cre mice. These mice were also crossed with Gnrh::gfp mice expressing green fluorescent protein under the control of the GnRH promoter in order to generate triple transgenic mice.
Isolation of GFP‐positive cells by FACS: schematic diagram showing the selection of the GFP‐positive population (green).
Relative mRNA expression from real‐time PCR analysis of the GnRH transcript in GFP‐positive cells, in comparison with GFP‐negative cells. The GnRH transcript appears to be selectively expressed in GFP‐positive cells. Mann–Whitney U‐test, n = 4 mice from 2 litters.
Relative mRNA expression from real‐time PCR analysis of neuropilin‐1 (Nrp1), Nrp2, plexin‐A1 (PlxnA1), and PlxnA4 transcripts in FACS‐isolated GFP‐positive GnRH neurons from control (Nrp1
loxP/loxP; gray) and mutant (Gnrh::cre; Nrp1
loxP/loxP; red) P0 mice. Unpaired t‐test, n = 5–6 mice per group from 3 litters.
Representative immunofluorescence images showing GnRH neurons migrating along Nrp1‐immunoreactive vomeronasal/terminal nerve fibers (empty arrowheads) at the nose–brain junction in sagittal slices from E14.5 control and mutant embryos. GnRH neurons (green) themselves express Nrp1 (red, white arrows) in control Nrp1
loxP/loxP mice, whereas Nrp1 is undetectable in GnRH neurons from mutant Gnrh::cre; Nrp1
loxP/loxP littermates (black arrows). Scale bar: 50 μm.
Data information: Bar graphs show individual values and means ± SEM. **
< 0.05.