Figure 2. Functional analysis of structural elements in Uba4.
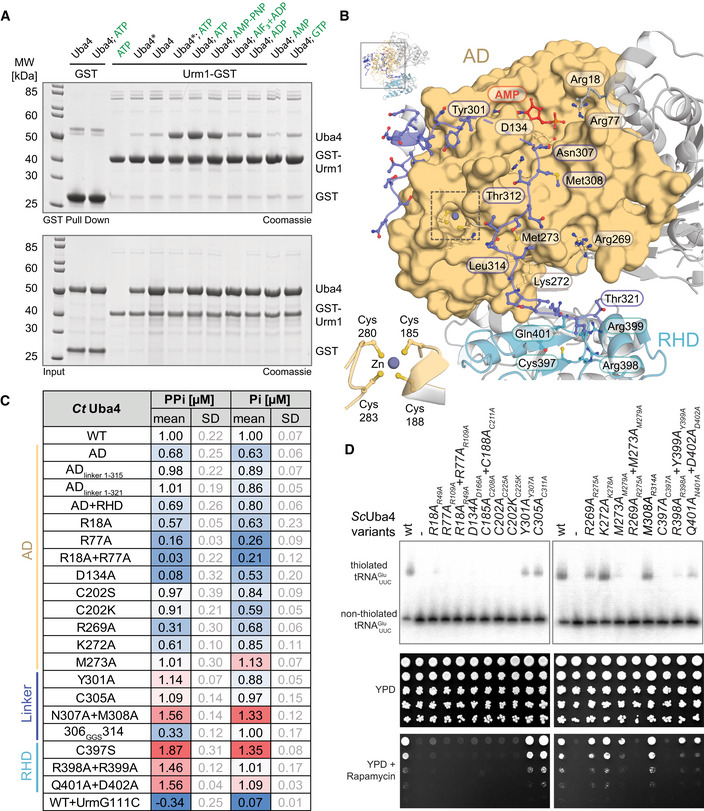
- Analysis of the interaction between CtUba4 and GST‐CtUrm1 (ratio 2:1 or 0.5:1 when marked with an asterisk) in the presence of nucleotide derivates by GST pull‐down resolved by SDS–PAGE and visualized with Coomassie stain.
- Close‐up (for reference see full structure on upper left) of the CtUba4 linker (aa 286–321) highlighting important residues and the coordination of the zinc site (inset, lower left).
- Tabular summary of ATP hydrolysis by CtUba4 WT and mutants in the presence of CtUrm1. The amount of released PPi was measured using a fluorometric pyrophosphate assay kit. After PPi hydrolysis, Pi was also detected by a Malachite Green kit. Changes in enzymatic activity are indicated as a color gradient from blue to red, normalized to CtUba4 WT. PPi: pyrophosphate; Pi: inorganic phosphate; SD: standard deviation; n ≥ 3.
- Functional analyses of ScUba4 mutant yeast using APM‐gel retardation (top) and viability in response to rapamycin (bottom). PAGE was supplemented with APM to retard the migration of thiolated tRNAs and allow their visualization by Northern blot using an anti‐ probe. APM: ([N‐Acryloyl‐amino]phenyl)mercuric chloride; YPD: yeast extract peptone dextrose. All residue numbering follows the CtUba4 sequence, but the respective ScUba4 numbering is added in subscript.
