Figure EV2. Structural details of apo Uba4.
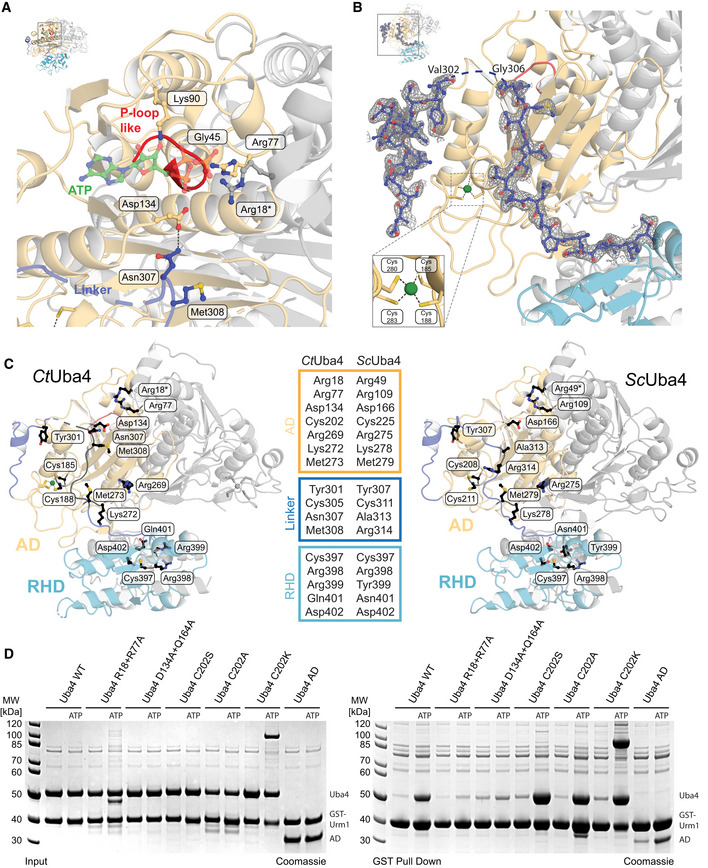
- Close‐up view of the CtUba4 P‐loop-like region and the nucleotide‐binding site. Amino acids required for ATP binding are shown as ball‐and-stick model colored according to atom type. The ATP molecule (green) was modeled based on the structural similarity to the ATP‐bound EcMoeB‐MoaD (PDB ID: 1JWA).
- Close‐up of the CtUba4 linker (aa 286–321) showing the refined 2F o‐F c electron‐density map contoured at 1.0σ. Coordination of the zinc site is highlighted.
- Cartoon representation of CtUba4 and the respective structural model of ScUba4 highlighting selected mutated amino acid residues shown as black ball‐and-stick model.
- Interaction analysis of WT and mutated CtUba4 with GST‐CtUrm1 by GST pull‐down in the absence and presence of 1 mM ATP. Input samples (left) and samples after GST pull‐down (right) are resolved by SDS–PAGE and visualized with Coomassie stain.
