Figure EV4. Structural comparison of the Uba4–Urm1 complex.
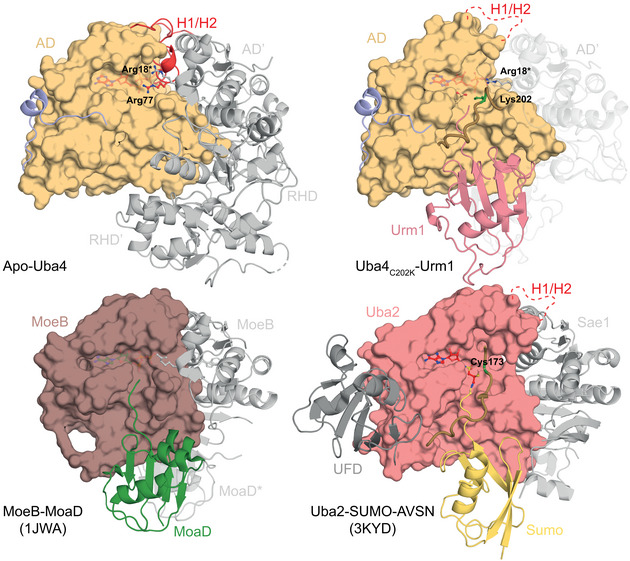
Comparison of the crystal structures of apo and Urm1‐bound CtUba4 (top) with the prokaryotic molybdenum‐cofactor‐biosynthesis protein MoeB bound to its substrate MoaD (PDB ID: 1JWA) and the eukaryotic SUMO E1 enzyme bound to its chemically modified substrate SUMO‐AVSN (PDB ID: 3KYD). E1‐like adenylation domains are shown as surface. The remaining domains or interacting proteins and bound UBL or SCP are shown as cartoon representation. The crossover loop containing the active cysteine (green) is highlighted. The H1/H2 helix that undergoes remodeling upon Urm1 modification is shown in red. The ATP molecule (red) is shown as ball‐and‐stick model.
