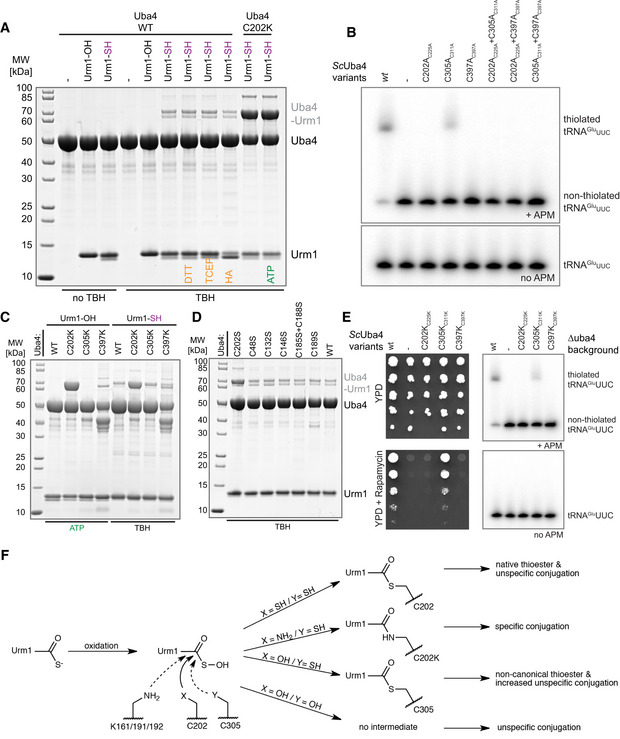
Analysis of covalent adduct formation between CtUba4 WT or C202K and carboxylated (–OH) or thiocarboxylated (–SH) CtUrm1 in the presence or absence of tert‐Butyl hydroperoxide (TBH). HA: Hydroxylamine; DTT: 1,4‐Dithiothreitol; TCEP: Tris(2‐carboxyethyl) phosphine; ATP: Adenosine triphosphate.
tRNA thiolation status of different ScUba4 mutant yeast strains analyzed by APM‐gel-retardation assay and Northern blot. APM: ([N‐Acryloyl‐amino]phenyl)mercuric chloride. All residue numbering follows the CtUba4 sequence, but the respective ScUba4 numbering is added in subscript.
Analysis of covalent adduct formation between WT or mutated CtUba4 and carboxylated (–OH) or thiocarboxylated (–SH) CtUrm1 in the presence of ATP and TBH, respectively.
Analysis of covalent adduct formation between WT or mutated CtUba4 and CtUrm1‐COSH in the presence of TBH.
Left: Viability analysis of ScUba4 mutant yeast in response to rapamycin. Right: Analysis of tRNA thiolation status in ScUba4 mutant yeast strains by APM‐gel-retardation assay and Northern blotting. Addition of APM to the gel retards the migration of thiolated tRNAs and allows their visualization. APM: ([N‐Acryloyl‐amino]phenyl)mercuric chloride. All residue numbering follows the CtUba4 sequence, but the respective ScUba4 numbering is added in subscript.
Schematic overview of various possible conjugation reaction routes of active‐site substitutions upon Urm1‐COSH oxidation. Main reaction path is indicated by a solid line and side reaction by a dashed line, respectively.