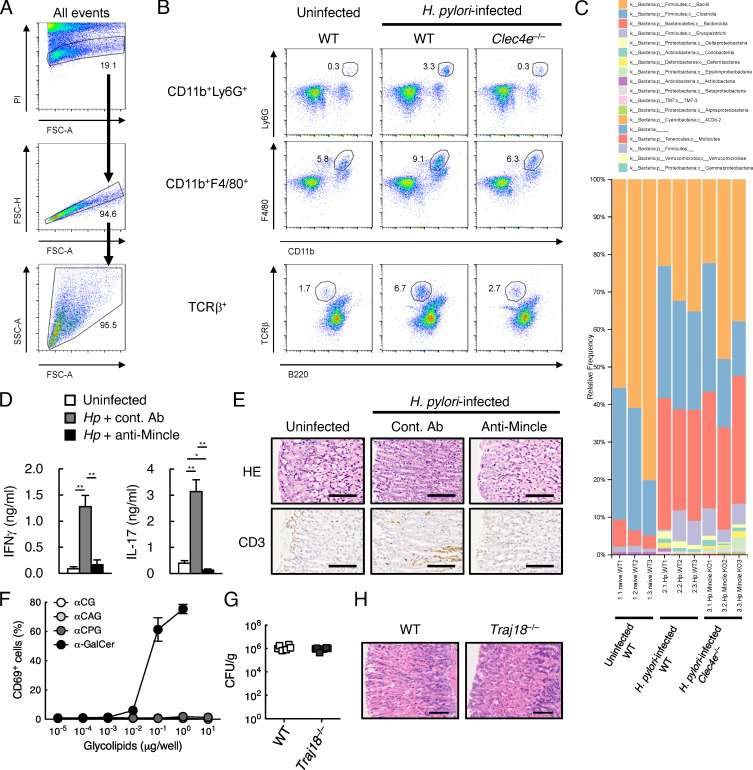
Figure S2.
Cellular immunity during H. pylori infection. (A) Gating strategy for analyzing gastric MNCs by flow cytometry. Numbers indicate the percentages of cells in each gate. PI, propidium iodide; SSC, side scatter; FSC, forward scatter. (B) Gastric MNCs from uninfected and H. pylori–infected WT or Clec4e−/− mice were stained with anti-Ly6G, F4/80, CD11b, TCRβ and B220 at 6 wk after infection. Numbers indicate the percentages of cells in each gate. (C) Metagenome analysis of gastric mucosal swabs from uninfected WT mice, H. pylori–infected WT and Clec4e−/− mice (n = 3 in each group) after 12 wk. (D) H. pylori–infected WT mice were injected with anti-Mincle mAb or rat IgG as a control Ab (cont. Ab). At 8 wk after infection, Peyer’s patch cells were collected and cultured for 4 d in the absence of exogenous H. pylori lysates. Uninfected WT mice were used as control. The concentrations of IFN-γ and IL-17 in the supernatants were determined by ELISA. (E) H&E (HE) staining and immunohistochemical staining with anti-CD3 of stomach sections from uninfected and H. pylori–infected mice treated with anti-Mincle mAb or control Ab. Scale bar, 100 µm. (F) Mouse invariant NKT hybridoma cells (DN32.D3) expressing CD1d were incubated with indicated amounts of αCG, αCAG, αCPG, or α-GalCer for 1 d. Each lipid was dissolved in DMSO. The expressions of CD69 were analyzed by flow cytometry. (G) The number of bacteria recovered from the stomachs of WT and Traj18−/− mice at 8 wk after H. pylori infection. Bacterial numbers were determined by counting the number of colonies on H. pylori selective agar plates. (H) H&E staining of stomach sections from H. pylori–infected WT and Traj18−/− mice after 8 wk. Scale bar, 100 µm. Data are presented as the mean ± SD of triplicate assays (D and F) from two independent experiments (at least six infected mice in each group) with similar results. An unpaired two-tailed Student’s t test was used for the statistical analyses. *, P < 0.05; **, P < 0.01.