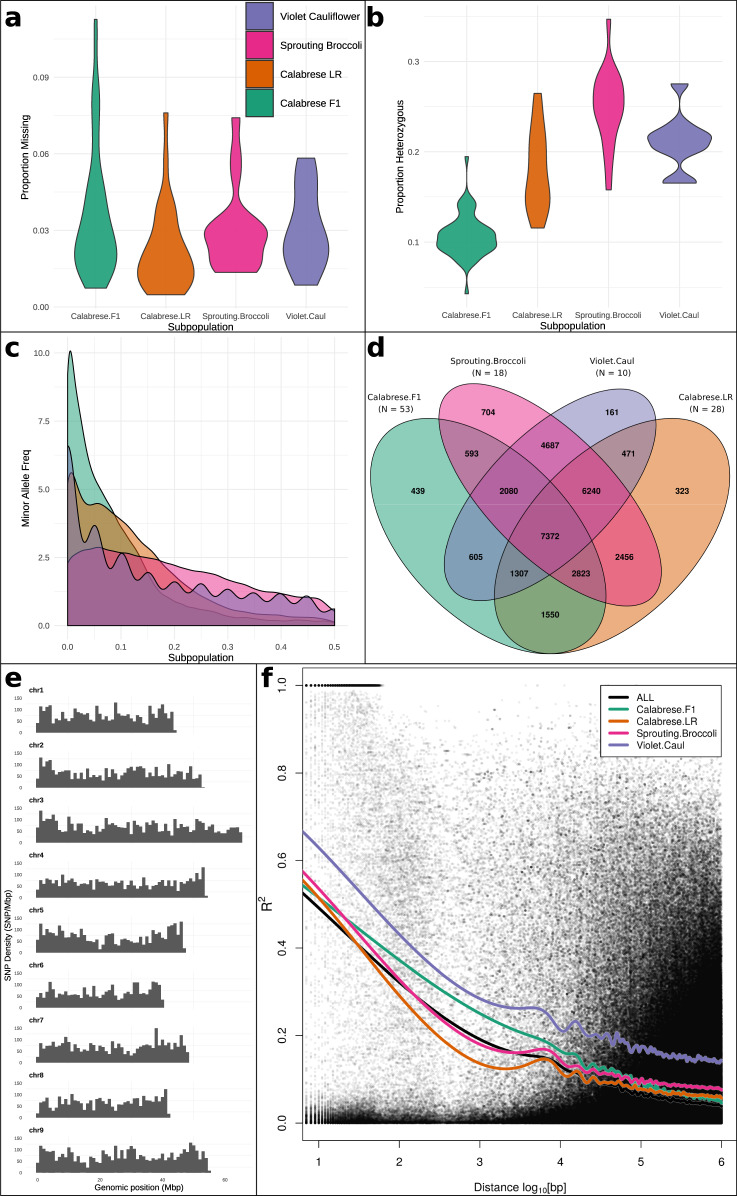
Fig. 2. GBS summary statistics for 31,811 high-quality SNP markers.
a Proportion of site missingness across all markers by subpopulation. b Proportion of site heterozygousity across all markers by subpopulation. c Pooled minor (second most common) allele frequency by subpopulation. d Unique polymorphic SNPs by subpopulation. e SNP density by chromosome (chr1-chr9) and genomic position in 1 Mbp windows (x-axis). f Linkage disequilibrium decay plotted as pairwise r2 against log10(bp) with subpopulation data fitted with cubic smoothing spline (spar = 0.5)