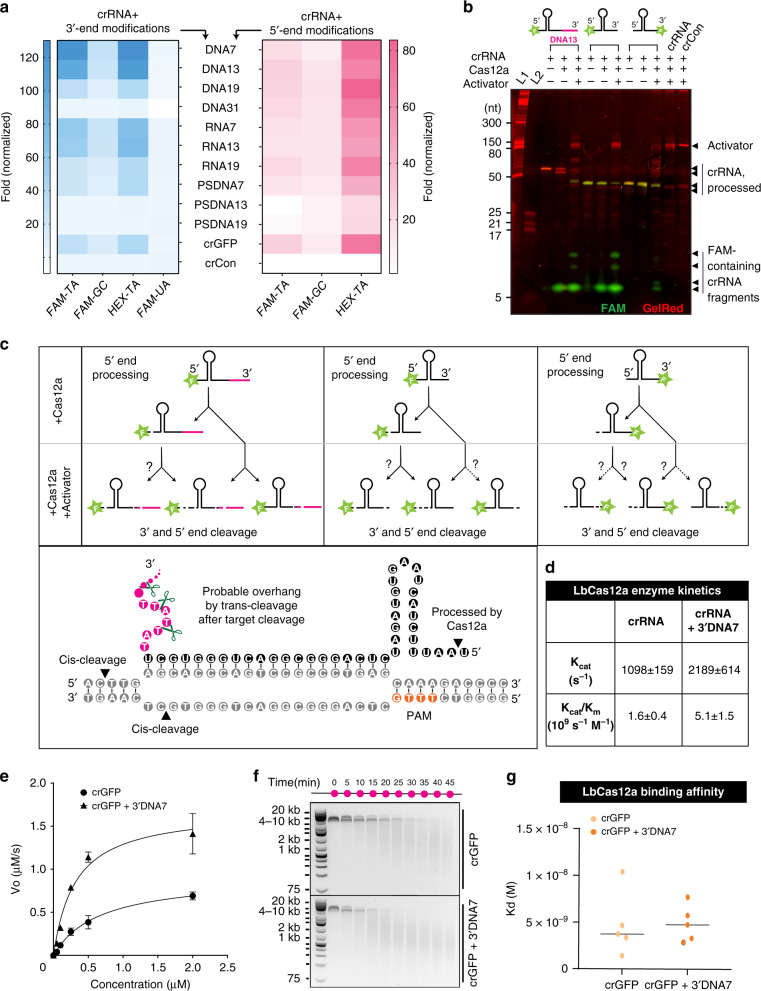
Fig. 2. Effect of reporter sequences on trans-cleavage activity, the proposed mechanism of crRNA end processing, enzyme kinetics, and binding affinity of LbCas12a with modified crGFP.
a Effect of different types of fluorophore-quencher systems on trans-cleavage activity with various modifications of crRNA. b Interactions of fluorescently labeled crRNAs with LbCas12a and dsDNA activator, characterized by PAGE analysis. In the absence of the activator, the modified crRNA (pre-crRNA) is trimmed by LbCas12a on its 5′-end (the first Uracil is cleaved, so-called truncated-crRNA or tru-crRNA). In the presence of the activator, the crRNA extensions are further trimmed, possibly leaving a 3′overhang. c Schematic diagram of putative processing of crRNA cleavage sites in the presence and absence of activator GFP. d Enzyme kinetic data of LbCas12a with crGFP vs. crGFP + 3′DNA7. e Michaelis-Menten kinetic study of the wild-type crGFP vs. crGFP + 3′DNA7. For this assay, 100 nM of LbCas12a, 100 nM of crRNA, and 7.4 nM of GFP fragment were used. Error bars represent mean ± SD, where n = 3 technical replicates. f Time-dependent cis-cleavage of LbCas12a on GFP in the presence of nonspecific ssDNA M13mp18. The reaction mixture was taken out every 5 min and quenched with 6× SDS-containing loading dye. g Dissociation constants of crGFP vs. crGFP + 3′DNA7. The Kd was determined by the biolayer interferometry binding kinetic assay with R2 > 0.9. Error bars represent ± SD, where n = 5 independent dilutions. Source data are available in the Source Data file.