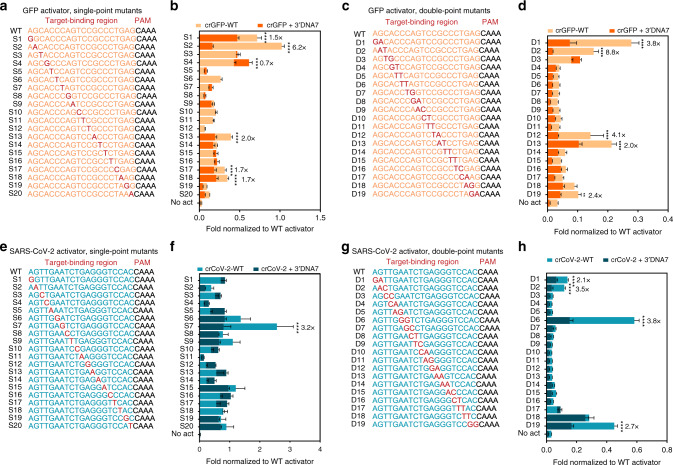
Fig. 4. Improved specificity of LbCas12a trans-cleavage with ENHANCE.
a Single-point mutations (S1-S20) and c double-point mutations (D1-D19) on the target strand of a dsDNA GFP activator. b Superimposed bar graphs indicating fold change in fluorescence of the S1-S20 mutant GFP activators normalized to the corresponding wild-type activator a. d Superimposed bar graphs indicating fold change in fluorescence of the D1-D19 mutant GFP activators normalized to the corresponding wild-type activator in c. e Single-point mutations (S1-S20) and g double-point mutations (D1-D19) on the target strand of a dsDNA SARS-CoV-2 activator. f Superimposed bar graphs indicating fold change in fluorescence of the S1-S20 mutant SARS-CoV-2 activators normalized to the corresponding wild-type (WT) activator in e. h Superimposed bar graphs indicating fold change in fluorescence of the D1-D19 mutant SARS-CoV-2 activators normalized to the corresponding wild-type activator in g. All the values b, d, f, and h were plotted after 20 min of incubation of various activators with wild-type CRISPR or ENHANCE. For b, n = 6 (three technical replicates examined over two independent experiments). For d, f, and h, n = 4 (two technical replicates examined over two independent experiments). For b, d, f, and h, values indicate mean ± SEM and the statistical analysis was performed using two-way ANOVA test with Dunnett’s multiple comparison test and only significant (p < 0.05) values were marked with an asterisk (*) indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. A fold change in specificity was calculated and reported for only statistically significant mutants by taking the ratio of the normalized data for crRNA-WT to crRNA-3′DNA7. Source data are available in the Source Data file.