Fig. 1 |. Workflow of single-cell RNA sequencing.
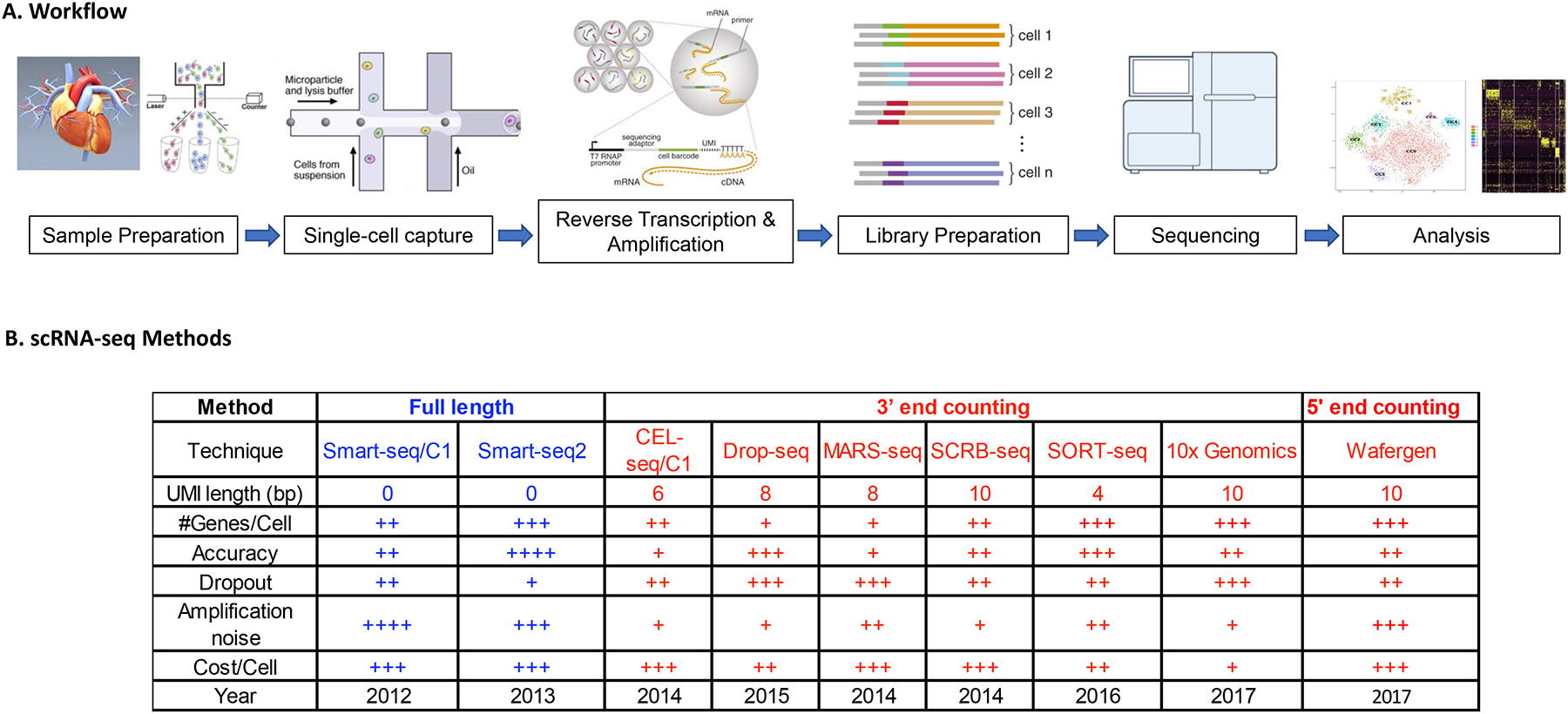
The general experimental workflow of single-cell RNA-sequencing begins with dissociation of the organ or tissue of interest to live single cells, which requires a fine-tuned digestion protocol that maximizes cell number and cell quality while minimizing the duration of digestion and cell death. Cultured cells are likewise detached and prepared as single cells. Prepared cells are then captured by various methods of single-cell capture. Reverse transcription of single-cell RNA is performed, followed by PCR amplification and library preparation of the resulting cDNA. Next-generation sequencing is subsequently performed to generate the readouts, which are aligned to a reference genome, processed for quality control and analysed by the user.
References for Fig. 1B
CEL-seq with UMI (Grün et al., 2014)
SCRB-seq (Soumillon et al., 2014)
MARS-seq (Jaitin et al., 2014)
STRT-C1 (Islam et al., 2014)
Drop-seq (Macosko et al., 2015)
CEL-seq2 (Hashimshony et al., 2016)
SORT-seq (Muraro et al., 2016)
DroNc-seq (Habib et al., 2017)
Seq-Well (Gierahn et al., 2017)
SPLiT-seq (Rosenberg et al., 2018)
sci-RNA-seq (Cao et al., 2017)
STRT-2i (Hochgerner et al., 2018)
Quartz-seq2 (Sasagawa et al., 2017)
10× Genomics Chromium (Zheng et al., 2017)
Wafergen ICELL8 (Gao et al., 2017)
Illumina ddSEQ SureCell
inDrops (Zilionis et al., 2017; Klein et al. 2015)
mcSCRB-seq (Bagnoli et al., 2018)
CEL-seq (Hashimshony et al., 2012)
Smart-seq (Ramskold et al., 2012)
Smart-seq2 (Picelli et al., 2013)
