Abstract
Antibody-oligonucleotide conjugates (AOCs) are a versatile class of chimeric biomolecules for therapeutics and biotechnological applications. Most widely employed chemical labeling methods for proteins are based on targeting of Lys or Cys residues that leads to mixed stoichiometry in the degree of conjugation and may interfere with antigen binding, thus, compromising the function of the antibody. A site-specific oligonucleotide conjugation technology providing full control over valency in mild reaction conditions would be an advancement to the state-of-the-art in bioconjugation. Herein, we demonstrate the production of single-chain variable fragment antibodies with fused SpyCatcher (scFv-SpyCatcher, monovalent) and alkaline phosphatase-SpyCatcher (scFv-AP-SpyCatcher, bivalent) on C-terminus and their conjugation to SpyTag002-oligonucleotide in phosphate-buffered saline (PBS). The formation of a covalent isopeptide bond between the protein and SpyTag002-oligonucleotide was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and the functionality of the obtained AOCs was confirmed in immuno-polymerase chain reaction (PCR) assays for the detection of microcystin-LR and 17β-estradiol. Based on time-resolved fluorescence immunoassays with scFv-AP fusion constructs, we observed that the SpyCatcher and SpyCatcher-SpyTag002-oligonucleotide part lowered the absolute signal obtained from the assay by 27.6 and 48.4% at 2 nM and by 26.2 and 27.6% at 100 pM microcystin-LR and 17β-estradiol concentrations, respectively. Nevertheless, the overall sensitivity of the immuno-PCR assays was similar to the time-resolved fluorescence immunoassays performed with the same components. In this study, vectors for SpyCatcher-fusion construction were created for directional cloning with SfiI sites enabling the rapid generation of AOC constructs for site-specific SpyTag-oligonucleotide conjugation.
1. Introduction
Antibody-oligonucleotide conjugates (AOCs) are a versatile class of chimeric biomolecules, which combine the unique functions of two fundamentally different types of biopolymers. The antibody part provides capacity for specific targeting of the epitope of interest, whereas the oligonucleotide part enables the implementation of antibody-oligonucleotide conjugates in a wide range of nucleic acid biochemistry-based applications. An application area of growing interest is the antibody-oligonucleotide conjugate-based pharmaceuticals, which primarily involve targeted antisense oligonucleotides or siRNA1,5 due to their potent and sustained gene silencing. Another major area of application is different detection approaches encompassing mainly immuno-polymerase chain reaction (PCR), proximity ligation/extension assays, electrochemical proximity assay, and DNA-PAINT imaging first and foremost by their exceptional sensitivity. The higher number of potential applications of antibody-oligonucleotide conjugates and antibody-oligonucleotide conjugation techniques have been recently reviewed by Dovgan et al.2
The production of a functional antibody-oligonucleotide conjugate requires the conjugation of the oligonucleotide(s) to the antibody firmly and without compromising the binding properties of the antibody. Currently, a vast majority of antibody conjugation techniques are based on targeting the nucleophilic primary amino group of lysine or the thiol group of reduced disulfides. However, a typical IgG molecule has around 80 Lys residues on its surface, with over 20 of them found at highly solvent-accessible sites3 resulting in a high level of nonspecific and stochastic labeling. All IgG molecules also contain several cysteines locked in structurally important disulfide bridges.4 Noncovalent approaches such as the streptavidin–biotin system or protein A/G utilization have also been devised for the generation of antibody-oligonucleotide conjugates.22,23 However, these methods also suffer from the lack of site-specificity, limited stability, and increased toxicity of the conjugates, e.g., in blood plasma.2,5 Better control over the number of oligonucleotides embedded within the antibody would improve the yield and help to manage the biophysical properties of antibody-oligonucleotide conjugates intended for either therapeutic or bioaffinity assay applications. The synthesis of such conjugates greatly benefits from a stable and site-specific labeling technique that can be achieved by incorporating peptide tags, e.g., Halo-tag, Sortase A, Avi biotin ligase recognition peptide, or modified amino acids.6 Other widely used site-specific conjugation strategies, such as GlyClick technology, involves the Fc domain of an antibody, making the conjugation of smaller antibody formats (Fab, scFv, diabody) impossible.
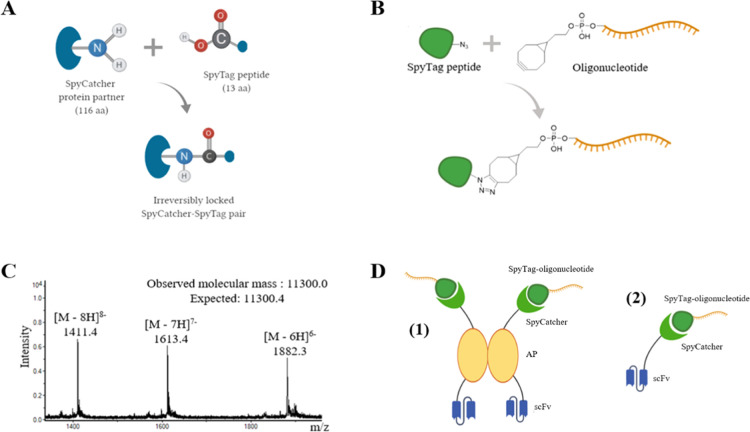
The unique protein ligation system based on splitting the second immunoglobulin-like collagen adhesin domain (CnaB2) from Streptococcus pyogenes into the 13-residue SpyTag peptide and the 116-residue SpyCatcher protein was discovered by Zakeri et al.8 The peptide, which corresponds to just one β-strand of the adhesion domain, forms a spontaneous amide bond with its protein partner (Figure 1A), enabling the protein–protein covalent conjugation without chemical cross-linking. The SpyCatcher-SpyTag system has proven to be a highly versatile tool for the construction of multifunctional biomolecular constructs as it has been successfully implemented, e.g., for modular assembly of proteins on nanoparticles,9 increasing thermostability of enzymes via circularization,10 labeling of cells for microscopic visualization,11 and even vaccine development.12
Figure 1.
(A) Isopeptide bond formation between SpyCatcher and SpyTag protein pair. (B) Synthesis of SpyTag-oligonucleotide conjugate; Strain-promoted alkyne-azide cycloaddition of the N-terminal azide to the bicyclononyne moiety in the 5′ end of the oligonucleotide. (C) Mass spectrometry (MS) electrospray ionization-time-of-flight (ESI-TOF) spectrum of the conjugate. Observed molecular mass is calculated from the most intense isotope combination at [(M – 6H)]6-/6. (D) Schematic representation of the dimeric (1) and monomeric (2) recombinants of the scFv-AP-SpyCatcher and scFv-SpyCatcher fusion proteins and its coupling with SpyTag-oligonucleotide.
Here, we report the development of an easy and efficient method for the production of antibody-oligonucleotide conjugates employing the SpyCatcher-SpyTag system as a tool for stable and site-specific bioconjugation. The method involves click chemistry-based synthesis of the SpyTag-oligonucleotide as well as expression of recombinant single-chain (scFv) antibody fragment as a fusion protein with SpyCatcher. Using this approach, we constructed antibody-oligonucleotide conjugates and analyzed their bifunctional properties, antigen recognition, and programmable specificity, with immuno-PCR and immunoassays in mono- and bivalent formats.
2. Results and Discussion
2.1. Synthesis of the SpyTag002-Oligonucleotide Conjugate and Generation of the AOCs
We set out to design a modular and robust platform for oligonucleotide conjugation to recombinant scFv antibodies using the SpyCatcher-SpyTag system, which allows the expression of the required fusion constructs in the bacterial host and the conjugation to take place at physiological pH. To this end, we constructed two expression vectors in which the scFvs are fused with either a C-terminal SpyCatcher or a C-terminal alkaline phosphatase AP-SpyCatcher fusion gene. Expression with the former vector, termed pHBSC, results in monomeric scFv-SpyCatcher fusion protein and with the latter, termed pLK06SC, in a dimeric scFv-AP-SpyCatcher construct13 with the added enzymatic reporter function of alkaline phosphatase (Figure 1D). The vectors were designed to be compatible with directional cloning with SfiI restriction sites enabling seamless conversion of scFvs from phage display selections or other in-house expression constructs14 into SpyCatcher fusions. To minimize the size of the fusion proteins, the core SpyCatcher sequence, 83 aa, was used in constructs.15 A flexible (G4S)3 linker was added between protein domains to allow independent domain folding and a His6 tag at the C-terminus for affinity purification.
The designed system was first tested by expressing the scFv NOD14 as a fusion to SpyCatcher and AP-SpyCatcher. NOD14 antibody (termed later as anti-microcystin-LR (MC-LR) scFv) specifically binds to the immunocomplex of the hepatotoxic cyclic peptide, microcystin-LR, and another antibody recognizing the ADDA residue ((2S,3S,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4(E),6(E)-dienoic acid) in the toxin.16,21 In our work, the expressed amounts of scFv-(AP)-SpyCatcher constructs were roughly 25–35 mg from 1 L of cell culture after purification. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis confirmed that scFv-SpyCatcher and scFv-AP-SpyCatcher protein bands appeared at the expected molecular weights, 39 and 89 kDa, respectively (Figure 2). The applicability of the system was also studied using another immunocomplex assay system intended for the detection of 17β-estradiol (E2), a steroid hormone essential for the development and sustaining of female reproductive tissues.17 To this end, the synthetic antibody library-derived scFv C618 (termed later as anti-E2 scFv), which binds to the immunocomplex of E2 and antibody S16, was cloned to pLK06SC vector, thus creating scFv-AP-SpyCatcher construct. The expression yield was similar to anti-MC-LR scFv-AP-SpyCatcher (Figure 2).
Figure 2.
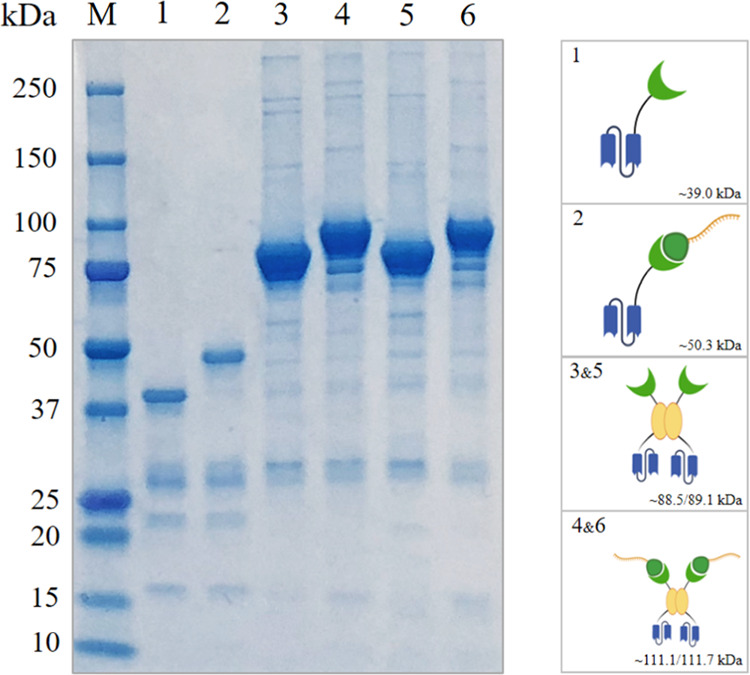
SDS-PAGE analysis of monomeric scFv-SpyCatcher (1), scFv-SpyCatcher-SpyTag002-oligonucleotide (2), and dimeric scFv-AP-SpyCatcher (3,5), scFv-AP-SpyCatcher-SpyTag002-oligonucleotide (4,6). M, molecular weight marker; lane 1, anti-MC-LR scFv-SpyCatcher; lane 2, anti-MC-LR scFv-SpyCatcher-SpyTag002-oligonucleotide; lane 3, anti-MC-LR scFv-AP-SpyCatcher; lane 4, anti-MC-LR scFv-AP-SpyCatcher-SpyTag002-oligonucleotide; lane 5, anti-E2 scFv-AP-SpyCatcher; lane 6, anti-E2 scFv-AP-SpyCatcher-SpyTag002-oligonucleotide. Abbreviations: scFv: single-chain variable fragment; MC-LR: microcystin-LR; AP: alkaline phosphatase; E2: 17β-estradiol.
SpyTag002 peptide,19 VPTIVMVDAYKPTK, used for the bioconjugation reaction with the generated scFv-(AP)-SpyCatcher constructs, shows six times faster association rate with SpyCatcher than the original SpyTag sequence. SpyTag002 peptide was conjugated with an ssDNA sequence using strain-promoted alkyne–azide cycloaddition (SPAAC), as it is fast and straightforward bioorthogonal reaction.20 The azide-modified SpyTag002 was assembled by an automated peptide synthesizer using the Fmoc/tBu–chemistry and commercially available amino acid building blocks. 2-(2-(2-azidoethoxy)ethoxy)acetic acid was coupled with the N-terminus to introduce the azide moiety. 2-Bicyclo[6.1.0]non-4-yn-9-yl (BCN)-modified oligonucleotide (5′-BCN-TTC ATC GCC CTT GCA AGC TTC GAC TAC CTA C-3′), in turn, was assembled using commercially available phosphoramidite building blocks. The SPAAC conjugation was performed on a 40 nmol scale by incubating these peptide/oligonucleotide-intermediates in 20 μL water for 6 h at 55 °C and exposing the crude product mixture directly to reversed phase high-performance liquid chromatography (RP HPLC). The desired peptide–oligonucleotide conjugate was obtained as the major product, and its authenticity was confirmed by mass spectrometry (Figure 1B).
The bioconjugation of the generated scFv-(AP)-SpyCatcher constructs and SpyTag002-oligonucleotide was performed in phosphate-buffered saline (PBS) (pH 7.4) for 12 h at 25 °C. The conjugates were purified with size exclusion chromatography (SEC) and analyzed with SDS-PAGE (Figure 2). As expected, the molecular size of the complexes increased by the mass of the SpyTag002-oligonucleotide (11.3 or 22.6 kDa in case of monomer and dimer, respectively).
2.2. Immuno-PCR for Microcystin-LR and 17β-Estradiol Detection
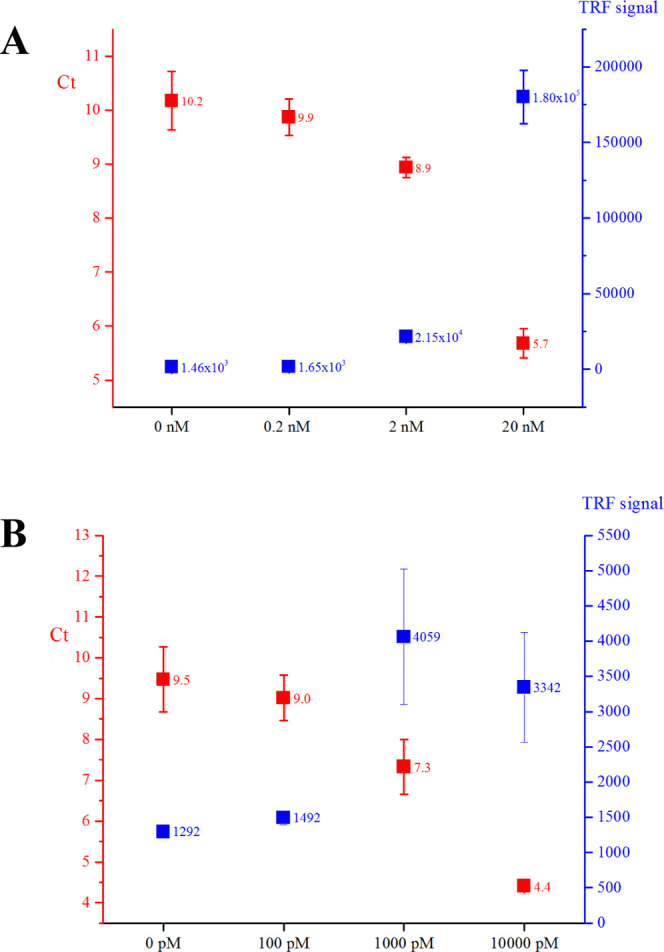
We evaluated the performance of the three different antibody-oligonucleotide conjugates (i.e., anti-MC-LR scFv-SpyCatcher-SpyTag002-oligonucleotide, anti-MC-LR scFv-AP-SpyCatcher-SpyTag002-oligonucleotide, and E2-scFv-AP-SpyCatcher-SpyTag002-oligonucleotide) in the immuno-PCR assay for the detection of low concentrations of MC-LR or E2. Briefly, magnetic microbeads were covered with capture antibodies following the addition of analyte and the appropriate immunocomplex binding scFv-oligonucleotide conjugate. After washing, the bound scFv-oligonucleotide conjugates were detected using the SYBR Green I-based real-time PCR amplification of the oligonucleotide sequence. Designed immuno-PCR for the detection of MC-LR showed a Ct (cycle threshold) dynamic range from 14.47 to 7.47 and from 9.87 to 5.68 for monomeric and dimeric constructs, respectively.
The Ct values obtained for the lowest MC-LR concentration (2 nM) were 12.75 ± 0.27 and 8.94 ± 0.19, for the monomeric and dimeric construct, respectively (Figure 3C). In both cases, a statistically significant difference, compared with the samples without MC-LR, was noticed. The observed results also show that the background level for monomeric and dimeric constructs is not the same: with the monomeric construct, the Ct value for the sample without MC-LR was 14.69 ± 0.26 vs 10.17 ± 0.54 with dimeric constructs. Moreover, Ct values for monomeric antibody-oligonucleotide conjugate were 1.3–1.5 times higher than those for a dimeric construct with each measured MC-LR concentration reflecting the presence of a greater oligonucleotide amount in the latter one.
Figure 3.
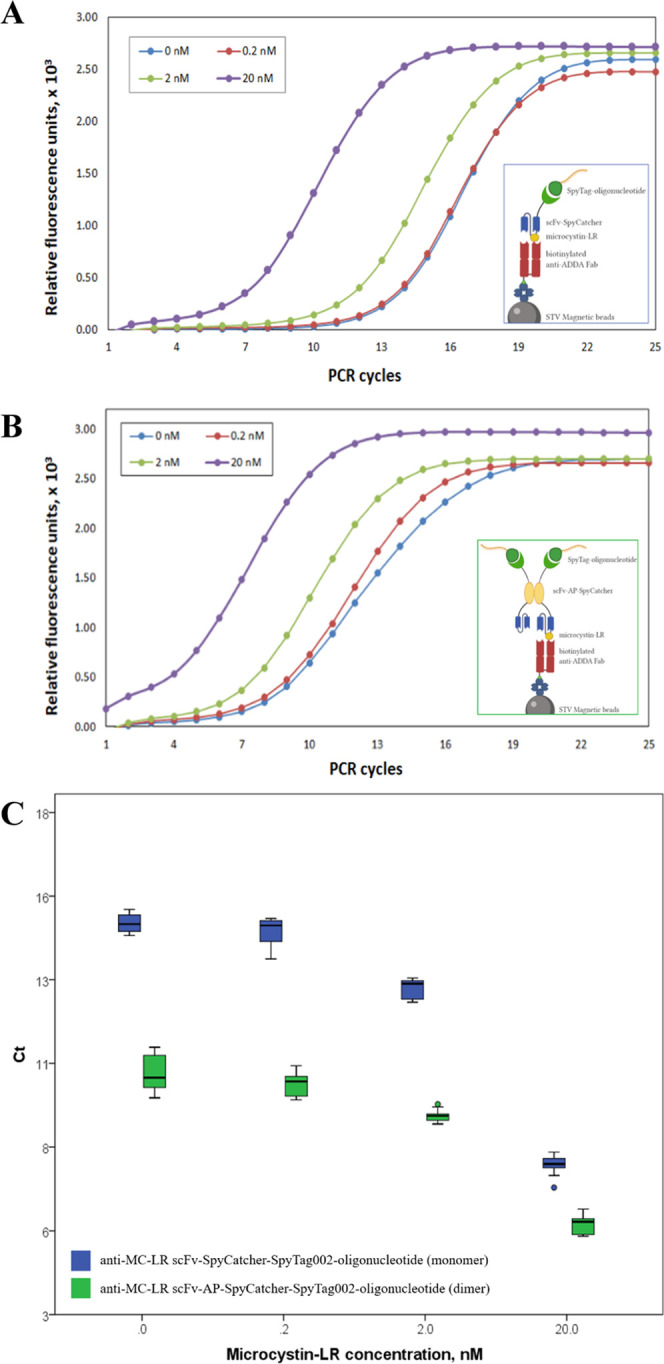
Representative results of real-time PCR for microcystin-LR detection. (A) Run profile vs PCR cycles. Lanes show the amplification profile of samples with different MC-LR concentrations ranging from 0 to 20 nM that have been detected with anti-MC-LR scFv-SpyCatcher-SpyTag002-oligonucleotide. (B) Run profile vs PCR cycles. Lanes show the amplification profile of samples with different MC-LR concentrations ranging from 0 to 20 nM that have been detected with anti-MC-LR scFv-AP-SpyCatcher-SpyTag002-oligonucleotide. (C) Threshold cycles (Ct) for MC-LR detection by different anti-MC-LR scFv constructs are shown; data are presented as mean ± standard deviation (SD) (n = 3).
For the detection of E2, five analyte concentrations between 1 and 10 000 pM were used, and the statistically significant difference in Ct was, at lowest, observed for 1000 pM E2 (Figure 5B, red boxes; Ct value 7.32 ± 0.67) in comparison to samples without E2 (marked as 0 pM; Ct value 9.47 ± 0.80).
Figure 5.
Comparison of the sensitivity of the immuno-PCR assay and the noncompetitive time-resolved fluorescence immunoassay for the detection of MC-LR (A) and E2 (B) with scFv-AP-SpyCatcher-SpyTag002-oligonucleotide constructs exploitation. Data are presented as mean ± SD (n = 3).
2.3. Performance of Obtained Conjugates in Time-Resolved Fluorescence Immunoassay (TRFIA)
To more closely assess whether the antibody-oligonucleotide conjugate construction-related modifications introduced into these antibodies affected their performance in immunoassay in general, the three different antibody constructs, scFv-AP, scFv-AP-SpyCatcher, and scFv-AP-SpyCatcher-SpyTag002-oligonucleotide, were tested side-by-side as detection antibodies in the TRF-immunoassays for both MC-LR and E2.
We observed a slight decrease in Eu signals for scFv-AP-SpyCatcher and scFv-AP-SpyCatcher-SpyTag002-oligonucleotide compared with the plain scFv-AP construct (Figure 4). The addition of the SpyCatcher domain to the fusion constructs lowered the absolute signal obtained from the assay by 27.6 and 26.2% at 2 nM and 100 pM MC-LR and E2 concentrations, respectively. The presence of oligonucleotide further decreased the signals by 20.8 and 1.4% at 2 nM and 100 pM MC-LR and E2 concentrations, respectively. This suggests that both the SpyCatcher and the oligonucleotide slightly weakens antibodies’ capacity to interact with the target immunocomplex. However, major sterical problems affecting the binding do not seem to exist, as expected due to the fusion of SpyCatcher to the C-terminus of the scFv, distant from the antigen-binding site. Alternatively, the added SpyCatcher and oligonucleotide can mask a part of the epitopes of the Eu-labeled anti-AP tracer antibody used in the assay. The compared constructs did not show major differences in terms of the unspecific binding, as indicated by the similar signal levels in the absence of the analyte (0 pM MC-LR or E2).
Figure 4.
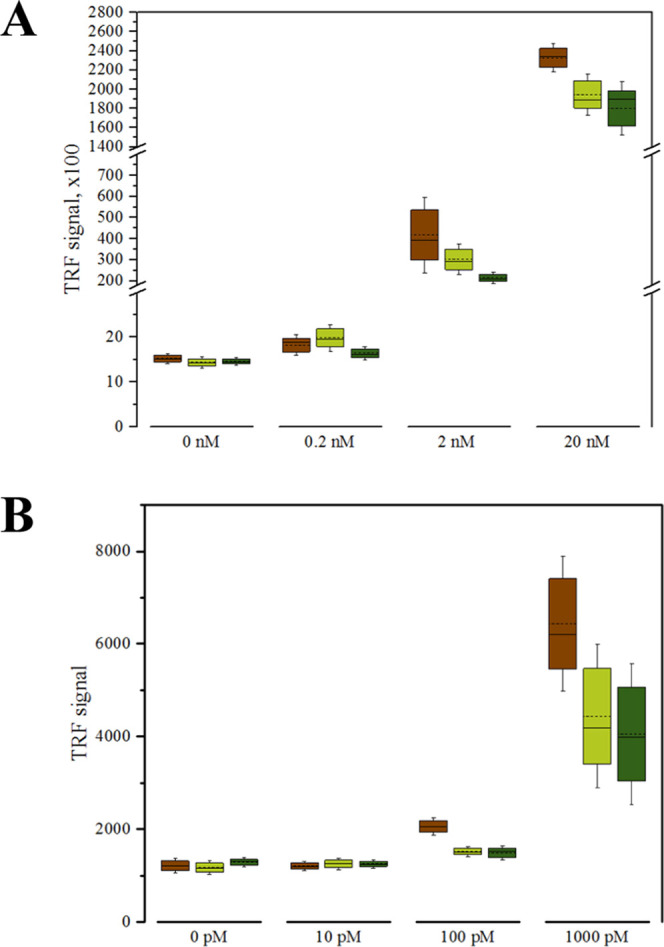
(A) Detection of microcystin-LR in the concentration range of 0.2–20 nM with scFv-AP construct (brown), scFv-AP-SpyCatcher (light green) and scFv-AP-SpyCatcher-SpyTag002-oligonucleotide (dark green). Data are presented as mean ± SD (n = 4). (B) Detection of E2 in the concentration range of 10–1000 pM with scFv-AP construct (brown), scFv-AP-SpyCatcher (light green), and scFv-AP-SpyCatcher-SpyTag002-oligonucleotide (dark green). Data are presented as mean ± SD (n = 3).
Altogether, the results of TRFIA also demonstrate that the performance of either of the scFvs is not severely compromised when converted to antibody-oligonucleotide conjugates.
The performance of scFv-AP-SpyCatcher-SpyTag002-oligonucleotide conjugates for the detection of MC-LR and E2 was evaluated in TRFIA vs immuno-PCR (Figure 5). In the case of MC-LR, the lowest analyte concentration yielding a statistically significant difference compared to the control was 0.2 nM in TRFIA, whereas the corresponding value in immuno-PCR was 2 nM (Figure 5A). For E2, the lowest concentration that could be detected with the scFv-AP-SpyCatcher-SpyTag002-oligonucleotide conjugate in TRFIA was 100 pM, while in immuno-PCR, the statistically significant difference was observed for 1000 pM, compared with the control (Figure 5B). In addition to the considerable background binding resulting in low Ct numbers even without analyte, the sensitivity of the immuno-PCR assay was affected by high intra-assay variation, especially at the lower analyte concentrations. A key to the improved immuno-PCR performance could be a more robust assay procedure (omitting the tedious manual handling of the microbeads used as solid phase) that could allow better control over the variation and reduction of the unspecific binding.
3. Conclusions
Antibody-oligonucleotide conjugates are versatile tools with a wide range of applications, both in vivo and in vitro. To facilitate the production of well-defined antibody-oligonucleotide conjugates, we report a convenient approach for the covalent site-specific attachment of a peptide-tagged oligonucleotide to a recombinantly produced scFv-SpyCatcher fusion protein in biocompatible conditions. Owing to the modular structure of an antibody, compact size of the SpyCatcher-SpyTag-system, and the well-established expression procedures for these recombinant proteins, the approach can be adapted to the production of different types of the antibody-oligonucleotide conjugate constructs, e.g., with different binding valencies.
The oligonucleotide conjugate of the relatively short SpyTag002-peptide (14 aa) can be prepared in a high yield using the SPAAC conjugation and the appropriate azide- and BCN-modified peptide, and oligonucleotide intermediates, which are needed for the conjugation, are readily accessible by automated synthesizers.
The functionality of antibody-oligonucleotide conjugates made with the developed approach was demonstrated using them to set up two immuno-PCR assays that allowed robust detection of the low-molecular-weight target analytes, MC-LR and E2. TRFIA experiments further confirmed the functionality of antibody-oligonucleotide conjugates; however, the results of TRFIA also implied that the addition of the SpyCatcher domain and the oligonucleotide could somewhat affect scFv’s capability to recognize the antigen (Figure 4). Theoretically, the immuno-PCR was expected to push the sensitivity down as compared to the TRFIA due to the virtually infinite specific response of the amplified signal. However, when compared using the same constructs (scFv-AP-SpyCatcher-SpyTag002-oligonucleotide), the observed sensitivities of immuno-PCR assays were similar to those of the TRFIA assays, and even somewhat weaker (Figure 5). The principal reasons for the limited sensitivity seem to lie in the high unspecific binding of the antibody-oligonucleotide conjugates to magnetic beads used as a solid phase in the immuno-PCR assays leading to the elevated background, i.e., low Ct-values without antigen (Figure 3). The other factor affecting the sensitivity of the immuno-PCR was high intra-assay variation. These limitations are also probably the reasons behind that the studied mono- and bivalent constructs, despite producing distinct Ct-value profiles, did not show a significant difference in the sensitivity of detection (Figure 3C). Optimization of the reaction conditions and/or switching the solid phase should allow further improving the sensitivity of the developed immuno-PCR assays.
The described approach is probably best suited to produce well-defined and stable antibody-oligonucleotide conjugates for various in vitro assay concepts since the bacterial origin of the SpyCatcher domain increases the risk of immunogenic reactions in the in vivo applications. However, the method may also find appropriate drug development implementation as an efficient way to produce multiple antibody-oligonucleotide combinations, with different binding specificities and/or nucleic acid cargo, for the early discovery stage cell-based studies.
4. Experimental Section
4.1. Materials and Equipment
All commercially available materials were used without further purification. For the immunoassays, all signal measurements were done with Victor 1420 multilabel counter (Wallac Oy, Finland). DELFIA wash and assay buffers, Europium fluorescence intensifier (EFI), and streptavidin-coated 96-well microtiter plates were purchased from Kaivogen Oy (Finland). Streptavidin-coated magnetic beads (MyOne C1 streptavidin-coated Dynabeads) were from Invitrogen (ThermoFisher Scientific). Anti-ADDA monoclonal antibodies, AD4G2, were from Enzo Life Sciences and were biotinylated using biotinisothiocyanate (BITC). The hormone, 17β-estradiol, was bought from Sigma-Aldrich. The BL21 and XL1 Escherichia coli cell lines used in the study were from Agilent Technologies. Cell lysates were prepared with Emulsiflex high-pressure homogenizer. Purification of scFv-AP-SpyCatcher and scFv-AP-SpyCatcher-SpyTag002-oligonucleotide fusion proteins was done with HisPur Ni-NTA resin (Thermo Scientific) and ÄKTA Prime Plus Purifier system (GE Healthcare Life Sciences, U.K.). The plasmid minipreps were done with the GeneJet plasmid miniprep kit (Thermo Scientific).
4.2. Synthesis of SpyTag002-Oligonucleotide and Production of scFv-(AP)-SpyCatcher for Microcystin-LR and 17β-Estradiol Detection
SpyTag002 peptide was assembled on a 10 μmol scale by an Applied Biosystems 433A peptide synthesizer using the Fmoc/tBu-chemistry and following routine procedures (PyBOP/DIEA was used in the coupling cycle). 2-(2-(2-Azidoethoxy)ethoxy)acetic acid was coupled into the N-terminus to introduce the azide moiety and the peptide was released by a mixture of anisol and trifluoroacetyl (TFA) (1:10, v/v, 2 h at RT). The crude peptide was precipitated in diethyl ether, purified by RP HPLC and lyophilized to dryness. bicyclo[6.1.0]nonyne (BCN)-modified oligonucleotide (5′-BCN-TTC ATC GCC CTT GCA AGC TTC GAC TAC CTA C-3′), initially analyzed for the absence of strong secondary structures with the program VectorNTI (ThermoFisher), was assembled on a 1 μmol scale by an Applied Biosystems 3400 DNA/RNA synthesizer using commercially available phosphoramidite building blocks. The oligonucleotide was released by concentrated ammonia (5 h at 55 °C), purified by RP HPLC, and lyophilized to dryness. The azide modified SpyTag002 peptide (40 nmol) and a slight excess of the BCN-modified oligonucleotide (46 nmol) were dissolved in water (20 μL) and they were allowed to react via strain-promoted alkyne-azide cycloaddition for 6 h at 55 °C. The mixture was purified by RP HPLC and lyophilized to dryness to yield 10 nmol of the conjugate (25%, determined by UV-absorption at λ = 260 nm). The authenticities of the peptide and oligonucleotide constituents and the product were verified by MS (ESI-TOF).
The anti-MC-LR and anti-E2 scFv genes were cloned to the pLK06H expression vector using SfiI restriction sites to produce the desirable antibody fragment as a fusion protein with SpyCatcher and with/without AP. Briefly, the plasmid was amplified in BL21-competent E. coli strain in SOC medium for 1 h (37 °C) and then was cultured on LA plates with 0.5% glucose and 50 μg/mL ampicillin overnight. Individual colonies from the transformation plate were cultured in 5 mL volume of SB medium supplemented with 50 μg/mL ampicillin and then were purified followed by the transformation into XL1-Blue-competent E. coli cells. For protein expression, transformed bacteria were grown in SB medium with 50 μg/mL ampicillin and 0.05% glucose to OD600 0.5–0.8 at 37 °C while shaking at 250 rpm. To induce protein expression, Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 200 μM and incubated for 12–14 h at 26 °C and 250 rpm. The cells were collected with 8000g centrifugation for 10 min at 4 °C and the obtained pellets were resuspended in the 200 mL volume buffer with the following content: 20 mM PBS, 0.5 kU nuclease, 1 mM MgCl2, and 10 mM imidazole. For purification, cells were lysed by two passages through an Emulsiflex C3 homogenizer and the obtained suspension was centrifuged twice at 4 °C for 30 min each time at 17 500g and 50 000g respectively. To purify the scFv-(AP)-SpyCatcher constructs, Ni-NTA affinity chromatography was applied followed by the purification with size-exclusion chromatography (SEC) to avoid dimers and tetramers. The desirable scFv-(AP)-SpyCatcher constructs obtained were easily proofed by SDS-PAGE (Figure 2).
Purified scFv-(AP)-SpyCatcher constructs were coupled to SpyTag002-oligonucleotide in 1:1 or 1:2 molar ratio in phosphate-buffered saline (PBS), pH 7.4, for 12 h at 25 °C for monomeric and dimeric scFv constructs, respectively. The coupled constructs were purified with SEC and checked in SDS-PAGE analysis (Figure 2).
4.3. Real-Time PCR Specificity and Sensitivity
Primer sequences were as follows: 003AKV_flank_fwd (5′-tgaaccagaggagttcttgcaTTCATCGCCCTTGCAAG-3′) and 004AKV_flank_rev (5′-ccagaatcacccgagcgaGTAGGTAGTCGAAGCTTGC-3′) for SpyTag002-oligonucleotide detection. The PCR mixtures were as follows: a reaction volume of 20 μL containing 2 μL of supernatant removed from beads, 0.5 μM of each primer, 1× Phusion HF reaction buffer, 0.2 mM dNTPs, 1× SYBR green, and 0.4 U of Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific). The PCR conditions were as follows: initial denaturation at 98 °C for 30 s, 40 cycles consisting of denaturation at 98 °C for 7 s, annealing at 60.6 °C for 10 s, and extension at 72 °C for 15 s, followed by a final extension at 72 °C for 180 s. For melting curve analysis, the PCR was followed by heating the reaction mixture from 65 to 95 °C with the 0.2 °C increments for 10 s each. To check original copy numbers per tube, the serial dilution of SpyTag002-oligonucleotide was made to the equivalent of 7.2 × 103, 7.2 × 105, 7.2 × 107, and 7.2 × 109 molecules per reaction. At least three replicates of SpyTag002-oligonucleotide were tested in each experiment. Nontemplate controls were also tested in triplicate using 2 μL of PCR-grade water. All samples were tested in triplicate, and results show the mean values generated unless otherwise stated. The curves of the amplification data (relative fluorescence units (RFU)) were analyzed by the software provided with the instrument, and the Ct values were defined as the number of cycles required for the fluorescent signal to cross the threshold.
4.4. Microcystin-LR and 17β-Estradiol Detection by Immuno-PCR
For the detection of microcystin-LR in immuno-PCR, 2 μL of magnetic beads per reaction were mixed with 0.8 μg of biotinylated anti-ADDA Fab in 100 μL volume and were incubated for 30 min at 23 °C on rotation. Then the beads were washed four times on magnet with 1× wash buffer. Serial dilutions of MC-LR, ranging from 0.2 to 20 nM, and Milli-Q ultrapure water (for blank samples) were added to the tubes together with scFv-SpyCatcher-SpyTag002-oligonucleotide or scFv-AP-SpyCatcher-SpyTag002-oligonucleotide to the final concentration 100 nM. After 1 h incubation time at 23 °C on rotation following four washing steps with 1× wash buffer, beads were resuspended in 20 μL of PCR water. Oligonucleotides from the beads were eluted by heating at 95 °C for 5 min. The supernatant was taken and used for real-time PCR as a template (2 μL/rxn). The real-time PCR steps were performed using the primers and reaction conditions described in Section 4.3. Each sample was performed in a triplicate. A melting curve analysis was carried out to evaluate the specificity of the test.
For the detection of 17β-Estradiol in immuno-PCR, 2 μL of magnetic beads per reaction were mixed with 0.8 μg of biotinylated anti-E2 S16 Fab7 in 100 μL volume and were incubated for 1 h at 23 °C on rotation. Then, the beads were blocked with 10 g/L bovine serum albumin dispersed in the assay buffer for 40 min at 37 °C. Blocked beads were washed four times on magnet with a 1× wash buffer. Then, E2 in a final concentration range from 10 to 10 000 pM or water (for blank samples) were added to the tubes together with scFv-AP-SpyCatcher-SpyTag002-oligonucleotide to the final concentration 100 nM. After 2 h of incubation time at 23 °C on rotation following four washing steps with 1× wash buffer, beads were resuspended in 20 μL of PCR water. Oligonucleotides from the beads were eluted by heating at 95 °C for 5 min. The supernatant was taken and used for real-time PCR as a template (2 μL/rxn). Each sample was performed in a triplicate. A melting curve analysis was carried out to evaluate the specificity of the test.
4.5. Microcystin-LR Detection in a Single-Step Noncompetitive Immunoassay
For immunoassay, the streptavidin-coated plates were washed four times before use and filled with MC-LR standards (final concentration range from 0.02 to 20 nM) in triplicates. Then, the reagent mixture was added to each well (biotinylated anti-ADDA Fab, 1 μg/μL; scFv desirable construct, 1 μg/μL; and N1–Eu-anti-AP pAb, 0.5 μg/mL) in 100 μL volume of the assay buffer. After 1 h incubation time at 23 °C on low shaking, plates were rewashed four times and the DELFIA enhancement solution was added to each well for 5 min. The TRF signal was measured with Victor 1420 fluorometer.
4.6. 17β-Estradiol Detection in Time-Resolved Fluorescence Immunoassay (TRFIA)
For immunoassay, the streptavidin-coated plates were washed four times before use and coated with 5 ng of biotinylated anti-E2 S16 Fab. After 1 h incubation at 23 °C on low shaking, plates were rewashed four times. Then, E2 in a final concentration ranging from 10 to 10 000 pM was added to the wells in a triplicate. On the top of the well, 10 ng of scFv-AP or scFv-AP-SpyCatcher or scFv-AP-SpyCatcher-SpyTag002-oligonucleotide were added to each well, and the plates were incubated for 2 h at 23 °C on low shaking. After four washing steps, the DELFIA enhancement solution was added to each well for 15 min. The TRF signal was measured with Victor 1420 fluorometer.
4.7. Statistical Analysis
All statistical analyses were performed with IBM SPSS Statistics 22.0, OriginLab 2016, and Microsoft Excel 2013.
Acknowledgments
This work was supported by Business Finland (New Modalities Ecosystem Project, 448/31/2018). Ms. Kushnarova-Vakal has financial support from the DPMLS (UTUGS, Finland).
Glossary
Abbreviations Used
- E2
17β-estradiol
- MC-LR
microcystin-LR
- Ct
cycle threshold
- AOC
antibody-oligonucleotide conjugate
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03750.
Detailed synthesis of azide-modified SpyTag002 peptide (S2), BCN-modified oligonucleotide (S5), and SpyTag002-oligonucleotide conjugate (S8). RP HPLC chromatogram of the azide-modified SpyTag002 peptide (Figure S1); structure and mass spectrum of SpyTag002 peptide (Figure S2); RP HPLC chromatogram of the BCN-modified oligonucleotide (Figure S3); structure and mass spectrum of the BCN-modified oligonucleotide (Figure S4); RP HPLC chromatograms of peptide–oligonucleotide conjugate (Figure S5); structure and mass spectrum of the peptide–oligonucleotide conjugate (Figure S6); SDS-PAGE analysis of the anti-MC-LR scFv-SC monomer (Figure S7); SDS-PAGE analysis of the anti-MC-LR scFv-SC-ST002-ON monomer (Figure S8); SDS-PAGE analysis of the anti-MC-LR scFv-AP-SC dimer (Figure S9); SDS-PAGE analysis of the anti-MC-LR scFv-AP-SC-ST002-ON dimer (Figure S10); SDS-PAGE analysis of the anti-E2 scFv-AP-SC dimer (Figure S11); and SDS-PAGE analysis of the anti-E2 scFv-AP-SC-ST002-ON dimer (Figure S12) (PDF)
Author Contributions
The manuscript was written through equal contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Huggins I. J.; Medina C. A.; Springer A. D.; van den Berg A.; Jadhav S.; Cui X.; Dowdy S. F. Site Selective Antibody-Oligonucleotide Conjugation via Microbial Transglutaminase. Molecules 2019, 24, 3287 10.3390/molecules24183287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovgan I.; Koniev O.; Kolodych S.; Wagner A. Antibody-oligonucleotide conjugates as therapeutic, imaging, and detection agents. Bioconjugate Chem. 2019, 30, 2483–2501. 10.1021/acs.bioconjchem.9b00306. [DOI] [PubMed] [Google Scholar]
- McCombs J. R.; Owen S. C. Antibody Drug Conjugates: Design and Selection of Linker, Payload and Conjugation Chemistry. AAPS J. 2015, 17, 339–351. 10.1208/s12248-014-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trads J. B.; Tørring T.; Gothelf K. V. Site-Selective Conjugation of Native Proteins with DNA. Acc. Chem. Res. 2017, 50, 1367–1374. 10.1021/acs.accounts.6b00618. [DOI] [PubMed] [Google Scholar]
- Leung D.; Wurst M. J.; Liu T.; Martinez R. M.; Datta-Mannan A.; Feng Yi. Antibody conjugates-recent advances and future innovations. Antibodies 2020, 19, 2 10.3390/antib9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. K.; El-Sayed A.; Barreto K.; Bernhard W.; Fonge H.; Geyer R. Site-specific fluorescent labeling of antibodies and diabodies using SpyTag/SpyCatcher system for in vivo optical imaging. Mol. Imaging Biol. 2019, 21, 54–66. 10.1007/s11307-018-1222-y. [DOI] [PubMed] [Google Scholar]
- Strop P.; Liu S. H.; Dorywalska M.; Delaria K.; Dushin R. G.; Tran T. T.; Ho W. H.; Farias S.; Casas M. G.; Abdiche Y.; Zhou D.; Chandrasekaran R.; Samain C.; Loo C.; Rossi A.; Rickert M.; Krimm S.; Wong T.; Chin S. M.; Yu J.; Dilley J.; Chaparro-Riggers J.; Filzen G. F.; O’Donnell C. J.; Wang F.; Myers J. S.; Pons J.; Shelton D. L.; Rajpal A. Location matters: site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem. Biol. 2013, 20, 161–167. 10.1016/j.chembiol.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Zakeri B.; Fierer J. O.; Celik E.; Chittock E. C.; Schwarz-Linek U.; Moy V. T.; Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E690–E697. 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.; Saccardo A.; Roccatano D.; Aboagye-Mensah D.; Alkaseem M.; Jewkes M.; Nezza F. D.; Baron M.; Soloviev M.; Ferrari E. Modular assembly of proteins on nanoparticles. Nat. Commun. 2018, 9, 1489 10.1038/s41467-018-03931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M.; Xu Q.; Jiang L.; Huang H. SpyTag/SpyCatcher cyclization enhances the thermostability of firefly luciferase. PLoS One 2016, 11, e0162318 10.1371/journal.pone.0162318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessino V.; Citron R.; Feng S.; Huang B. Covalent protein labeling by SpyTag-SpyCatcher in fixed cells for super-resolution microscopy. ChemBioChem 2017, 18, 1492–1495. 10.1002/cbic.201700177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune K. D.; Buldun C. M.; Li Yu.; Taylor I. J.; Brod F.; Biswas S.; Howarth M. Dual plug-and-display synthetic assembly using orthogonal reactive proteins for twin antigen immunization. Bioconjugate Chem. 2017, 28, 1544–1551. 10.1021/acs.bioconjchem.7b00174. [DOI] [PubMed] [Google Scholar]
- Suzuki C.; Ueda H.; Suzuki E.; Nagamune T. Construction, bacterial expression, and characterization of hapten-specific single-chain Fv and alkaline phosphatase fusion protein. J. Biochem. 1997, 122, 322–329. 10.1093/oxfordjournals.jbchem.a021756. [DOI] [PubMed] [Google Scholar]
- Huovinen T.; Syrjanpaa M.; Sanmark H.; Brockmann E.-C.; Azhayev A.; Wang Q.; Vehniäinen M.; Lamminmäki U. Two scFv antibody libraries derived from identical VL-VH framework with different binding site designs display distinct binding profiles. Protein Eng., Des. Sel. 2013, 26, 683–693. 10.1093/protein/gzt037. [DOI] [PubMed] [Google Scholar]
- Li L.; Fierer J. O.; Rapoport T. A.; Howarth M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide tag. J. Mol. Biol. 2014, 426, 309–317. 10.1016/j.jmb.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi S.; Kato H.; Mizuno M.; Tsiji K.; Harada K. Bacterial degradation of microcystins and nodularin. Chem. Res. Toxicol. 2005, 18, 591–598. 10.1021/tx049677g. [DOI] [PubMed] [Google Scholar]
- Dai Y.; Liu C. C. Detection of 17β-estradiol in environmental samples and for health care using a single-use, cost-effective biosensor based on differential pulse voltammetry (DPV). Biosensors 2017, 7, 15 10.3390/bios7020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivo J.; Kivimäki L.; Juntunen E.; Pettersson K.; Lamminmäki U. Development of anti-immunocomplex specific antibodies and non-competitive time-resolved fluorescence immunoassay for the detection of estradiol. Anal. Bioanal. Chem. 2019, 411, 5633–5639. 10.1007/s00216-019-01952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble A. H.; Banerjee A.; Ferla M. P.; Reddington S. C.; Anuar I. N. A. K.; Howarth M. Evolving accelerated amidation by SpyTag/SpyCatcher to analyze membrane dynamics. Angew. Chem., Int. Ed. 2017, 56, 16521–16525. 10.1002/anie.201707623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- Akter S.; Vehniäinen M.; Spoof L.; Nybom S.; Meriluoto J.; Lamminmäki U. Broad-spectrum noncompetitive immunocomplex immunoassay for cyanobacterial peptide hepatotoxins (Mycrocystins and Nodularins). Anal. Chem. 2016, 88, 10080–10087. 10.1021/acs.analchem.6b02470. [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M.; Boldt L.; Ceyhan B.; Blohm D. DNA-Directed immobilization: efficient, reversible, and site-selective surface binding of proteins by means of covalent DNA-streptavidin conjugates. Anal. Biochem. 1999, 268, 54–63. 10.1006/abio.1998.3017. [DOI] [PubMed] [Google Scholar]
- Jung Y.; Lee J. M.; Jung H.; Chung B. H. Self-directed and self-oriented immobilization of antibody by Protein G-DNA conjugate. Anal. Chem. 2007, 79, 6534–6541. 10.1021/ac070484i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.