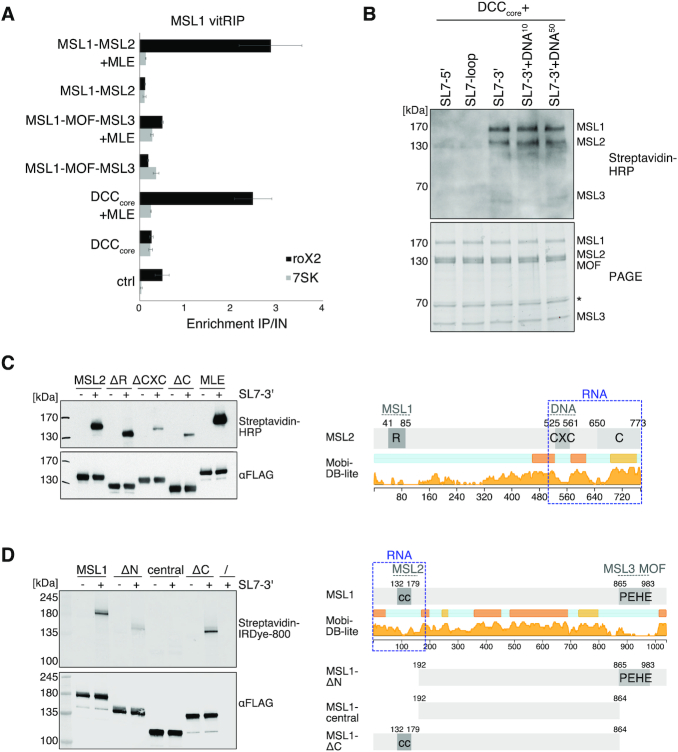
Figure 5.
The MSL1-MSL2 module of the DCC recruits MLE and incorporates remodeled roX2. (A) vitRIP of recombinant DCC DNA-binding module (MSL1-MSL2), reader-writer module (MSL1-MOF-MSL3) and DCCcore (MSL1-MSL2-MOF-MSL3) with S2 RNA in absence or presence of [Low] MLE and ATP. vitRIP utilized an MSL1 antibody. Enrichment of roX2 and 7SK was analyzed by RT-qPCR and is displayed relative to input. Ctrl indicates an RNA-only MSL1 vitRIP sample lacking recombinant proteins. Error bars represent standard deviation for two or three independent replicates. Statistical analysis of RT-qPCR is given in Supplementary Table S3. (B) UV crosslinking assay showing direct binding of the DCCcore to biotinylated roX2 RNA fragments mimicking alternative secondary structures. Unwinding of stem loop 7 (SL7) in the 3′ region of roX2 by MLE generates alternative stem loops as predicted in Supplementary Figure 9B. Annotation of roX2 secondary structure is based on (25). The presence of 10 pmol or 50 pmol PionX CG8097 dsDNA in addition to 50 pmol RNA in the reaction is indicated by DNA10 and DNA50, respectively. Protein-crosslinked biotinylated roX2 fragments were detected by Streptavidin-HRP (upper panel). The DCCcore proteins were visualized in a stain-free image of the SDS-PAGE (lower panel). The asterisk indicates a co-purifying contaminant. (C) Left: UV crosslinking assay showing direct binding of 10 pmol MLE and MSL2 to 50 pmol biotinylated remodeled roX2 SL7-3′ RNA. Of MSL2, wild-type and the mutants ΔRING (MSL2Δ1-131), ΔCXC (MSL2Δ523-565) and ΔC (MSL2Δ650-778) were analyzed. Upper panel shows the protein-bound biotinylated SL7-3′ RNA detected by Streptavidin-HRP. Lower panel shows an anti-FLAG western blot for detection of FLAG-tagged MLE and MSL2 proteins, respectively. Right: Domain structure of MSL2. The RING (R), CXC, and C-terminal domain (C) are labeled. Intrinsic disorder probability was predicted by MobiDB 3.0 (72). Dashed gray lines indicate known DCC interfaces. Dashed blue box indicates the putative non-canonical RNA binding region in MSL2. (D) UV crosslinking assay probing 10 pmol MSL1 for direct binding to 50 pmol biotinylated remodeled roX2 SL7-3′ RNA. Of MSL1, wild-type and the mutants ΔN (MSL1Δ1-191), central (MSL1192-864) and ΔC (MSL1Δ865-1039) were analyzed. Protein-bound biotinylated SL7-3′ RNA was detected using IRDye 800 Streptavidin. FLAG-tagged MSL1 variants were detected by anti-FLAG western blot. Right: Domain structure of MSL1. The coiled-coil region (cc) and the PEHE domain are indicated. Intrinsic disorder probability was predicted by MobiDB 3.0 (72). Dashed gray lines indicate known DCC interfaces. Dashed blue box indicates the putative non-canonical RNA binding regions in MSL1.