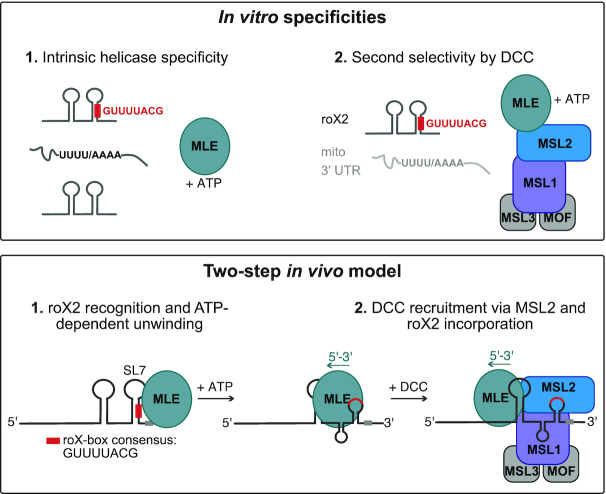
Figure 6.
Two-step model for selective roX2 integration into the DCC. Upper panel: In vitro, the helicase MLE exhibits intrinsic specificity for U-/A-rich linear sequence motifs and tandem stem-loop structures. These features are combined in roX1/2 RNA. Here the stems of RNA secondary structures contain stretches of U:A base pairing. ATP-dependent unwinding thus exposes linear U-/A-rich sequences. The DCCcore provides additional selectivity, which increases the affinity to the physiological substrate roX2 and decreases the affinity to other U-/A-rich transcripts. Lower panel: In the first step of roX2 integration into the DCC, MLE uses its intrinsic specificity to bind and unwind roX2 with 3′–5′ directionality. The unwinding process exposes binding sites on the RNA (the conserved roX-box consensus sequence). In the second step, additional selectivity provided by the DCCcore prevents unspecific binding of other MLE substrates and allows specific interaction of MLE and DCCcore via MSL2. Within the DCC, the MSL1-MSL2 module contributes to roX2 integration by direct binding to exposed roX-box sequences.