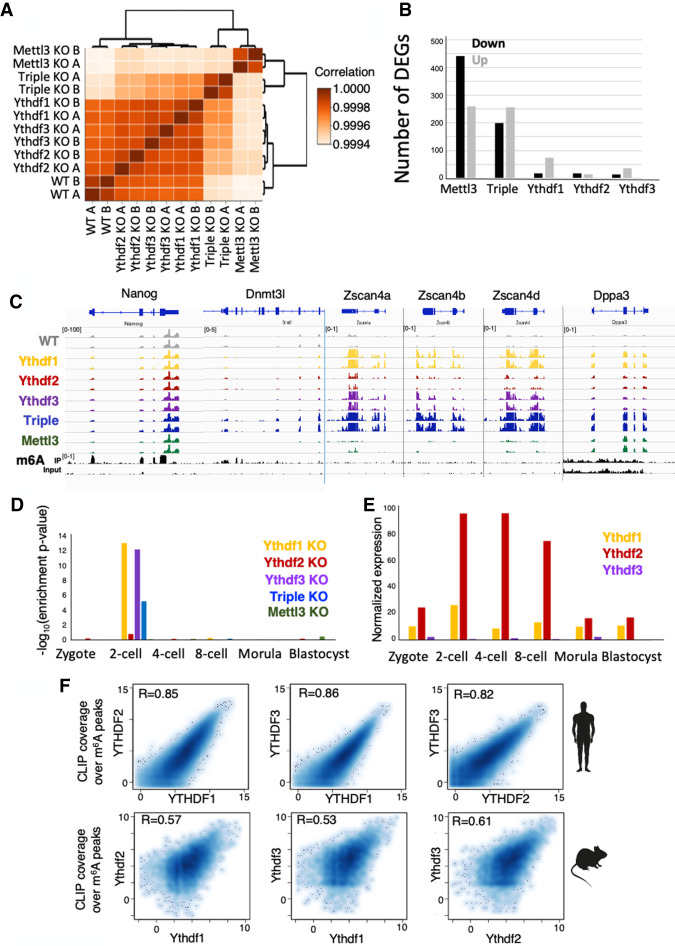
Figure 6.
Ythdf triple-KO has a dramatic effect on gene expression. (A) Hierarchical clustering of samples based on Pearson correlation, showing that single-KO samples are highly similar to WT. (B) Number of differentially expressed genes in each of the KO cell lines, compared with WT. (Black) Down-regulated genes, (gray) up-regulated genes. (C) RNA-seq and m6A methylation landscape of selected genes. Normalized coverage is presented. Only Nanog and Dnmt3l are m6A methylated. Dnmt3l, Zscan4a, Zscan4b, Zscan4d, and Dppa3 are overexpressed in triple-KO. (D) Enrichment of up-regulated genes in each category, to early embryo genes (Gao et al. 2017). Genes that are up-regulated in KO of Ythdf1 and Ythdf3 are specifically enriched for two-cell embryo genes. (E) Normalized expression of Ythdf1, Ythdf2, and Ythdf3, as measured in early mouse embryo (Gao et al. 2017). (F) CLIP coverage over m6A peaks of Ythdf readers as measured in humans (n = 41,885) (Patil et al. 2016), and in mice (n = 9861), showing high correlation between coverage by different Ythdf readers.