Figure 1.
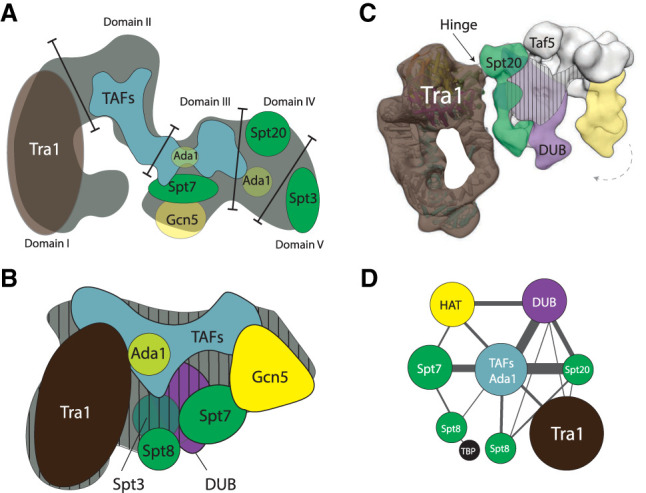
Schematic overview of the architecture of the SAGA complex based on electron microscopy, immunolabeling, and cross-linking mass spectrometry techniques. (A) The first S. cerevisiae SAGA structure divides it in five domains (adapted from Wu et al. 2004, with permission from Elsevier). Tra1 occupies domain I, and domains II–IV contain TAFs, core (Ada1, Spt7, and Spt20) and HAT subunits (Gcn5). Domain V is flexible and contains Spt3. (B) The subsequent SAGA structure locates the DUB module as bulging density in close proximity to TAF and Spt subunits. (Adapted from research originally published in Setiaputra et al. 2015. © the American Society for Biochemistry and Molecular Biology). This structure highly resembles the previous structure (Wu et al. 2004), when domain V is flipped toward domain I. The Spt subunits form a TBP-binding surface adjacent to Tra1, and the DUB and HAT modules locate to the right. Vertical dashes indicate the midplane. Subunit Spt3 and the DUB module are located on the backside. (C) Cryo-EM structure showing the narrow hinge region that forms the peripheral connection between Tra1 and the core modules (arrow; Tra1 cross-links to Spt20 and Taf12). The DUB (purple) and HAT module (yellow) associate with the structural core (green/white). The vertical stripes indicate the midplane. The DUB module anchors via the backside. Both enzymatic modules form a clamp that engages nucleosomes, and the HAT module can swirl ∼15° toward the DUB module (dashed arrow) (adapted from Cheung and Díaz-Santín 2019). (D) The architectural map determined by cross-linking mass spectrometry (adapted with permission from Han et al. 2014; © 2014 The Authors) reveals the close proximity of Tra1 and the Spt subunits, and the peripheral connections of Tra1 to the rest of the complex. The TAFs with Ada1 assume a central position and are interlinked with Spt subunits. The HAT and DUB module link to the TAF/Ada1 center, and the DUB module cross-links also to Spt20.
