Figure 3.
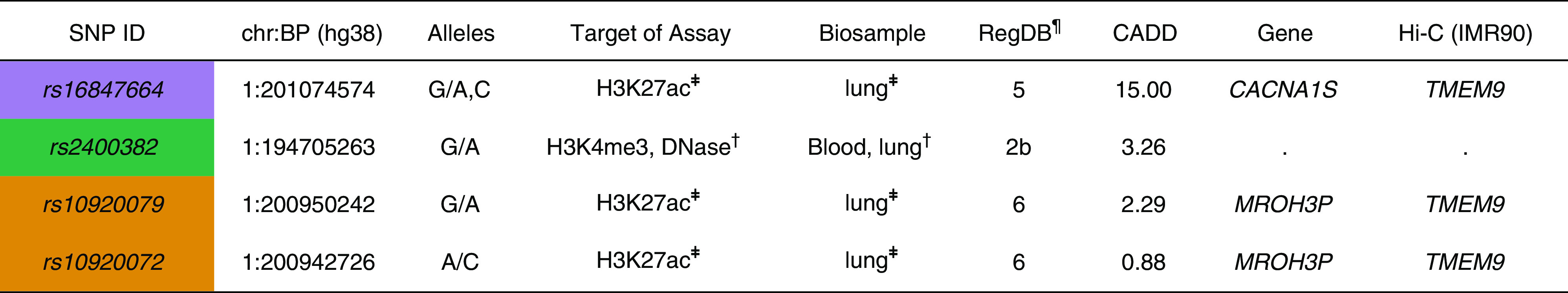
Functional annotation of four variants identified from fine-mapping analysis at 1q32. Each color represents SNPs identified from fine-mapping conditional analysis and their linkage disequilibrium SNPs (R2 > 0.8) with at least one or more than one regulatory element data. ¶RegulomeDB (RegDB) score represents the likelihood that a variant will be located within a functional region. The RegDB variant classification scheme is defined as follows (50): Likely to affect binding: 2b, TF binding + matched TF motif + matched DNase footprint + DNase peak. Minimal binding evidence: 4, TF binding + DNase peak; 5, TF binding or DNase peak; 6, motif hit. ‡Overlapping with H3K27ac chromatin immunoprecipitation sequencing peaks in human bronchial epithelial cells. †High signals of epigenetic markers in immune cells and lung-related tissues (Encyclopedia of DNA Elements data). BP = base pair position; CADD = Combined Annotation-Dependent Depletion; chr = chromosome; Hi-C = high-throughput chromosome conformation capture; TF = transcription factor.
