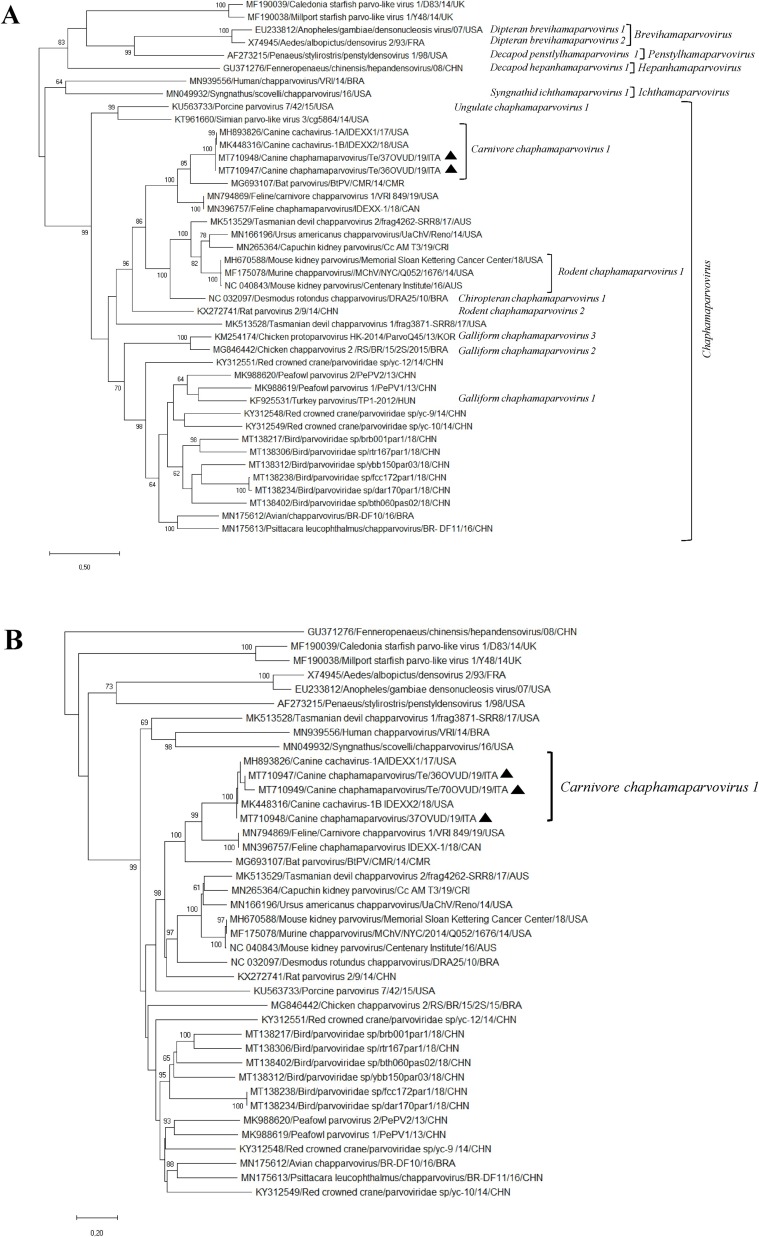
Fig. 1.
Phylogenetic analyses based on the aa sequence of the NS1 (A) and VP (B) of the CaChPVs identified in this study. The trees, constructed with a selection of ChPV strains representative of each species, were generated using Maximum Likelihood method based on the Poisson correction and supplying statistical support with bootstrapping of 1000 replicates. The scale bar indicates nucleotide substitutions per site. Black triangles indicate the CaChPV strains detected in this study. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).