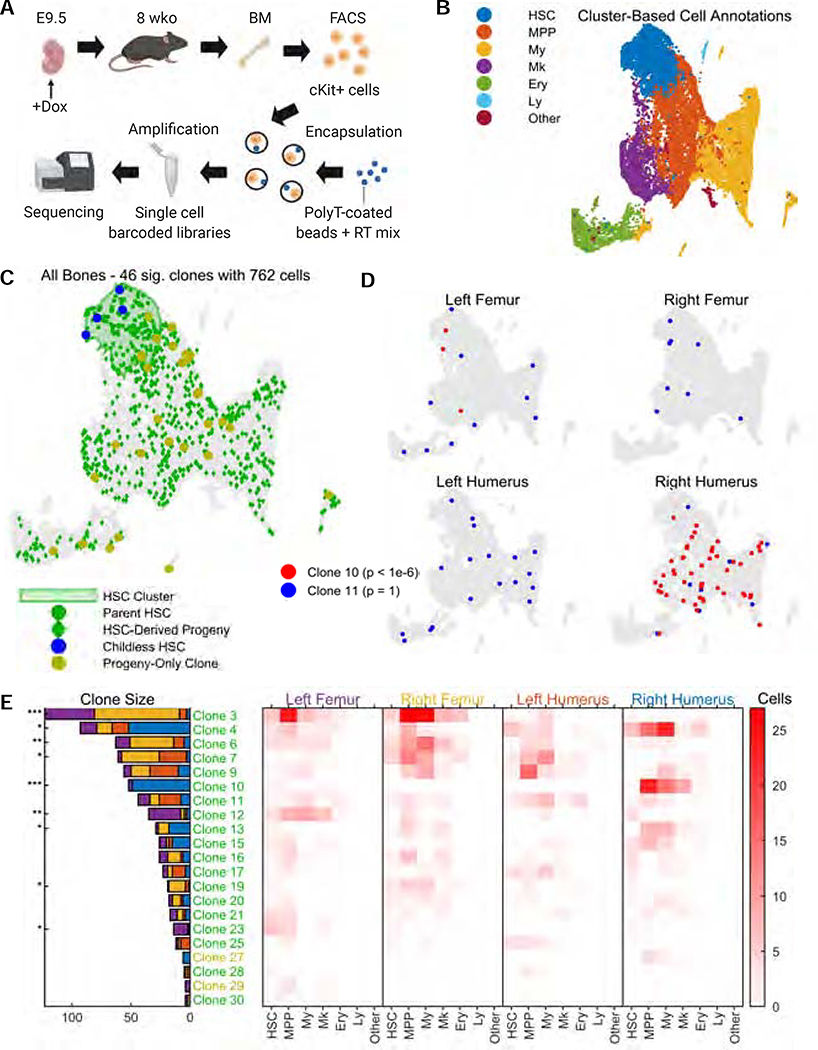
Figure 5: Clonal tracing of blood progenitors to adulthood.
A. CARLIN mice were labelled at E9.5. At 8 weeks, bone marrow cells were collected, sorted, and encapsulated for single-cell RNA sequencing. Schematic created with BioRender.
B. UMAP representation of pooled transcriptome data from the bone marrow of 4 separate bones. See Supplementary Figure 5D,E for the breakdown of clusters and markers used for annotation. HSC, hematopoietic stem cell; MPP, multipotent progenitor cell; My, myeloid progenitor cells; Ery, erythrocyte; Ly, lymphoid cell.
C. Statistically significant CARLIN alleles (FDR = 0.05; Methods) across all bones combined, overlaid onto the UMAP plot from (B). The green shaded area corresponds to the HSC cluster in the transcriptome, shown in (B). We are able to directly map the ancestry between differentiated cells (green diamonds) and HSCs (green circles) which share the same set of alleles. HSCs without children are shown in blue, and differentiated cells that do not share their allele with HSCs are shown in yellow.
D. CARLIN clones overlaid onto the transcriptome of individual bones; a non-biased clone (blue) and a biased clone (red) are shown with the Bonferroni-corrected p-values for bone bias (Methods).
E. (Left) Bar graph indicating the prevalence of each statistically significant allele across the 4 bones, with the Bonferroni-corrected p-value for bone bias marked as *p<0.05; **p<10−3; ***p<10−6. (Right) Heatmap indicating occurrence frequency of alleles across bones and cell types. Alleles found in fewer than 4 cells are not displayed. The clone labels follow the color scheme in (C).