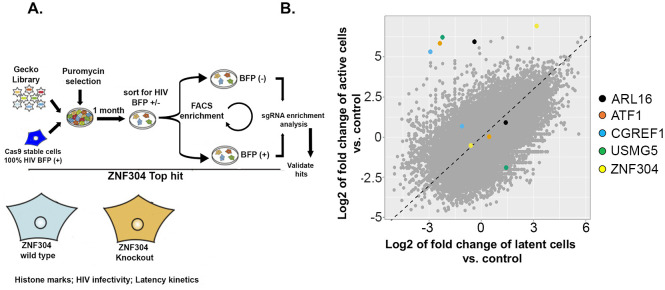
Fig 1. Genome-wide CRISPR knockout screen for identifying host factors that promote HIV latency.
A. Schematic representation of the genome CRISPR KO screen towards identifying host factors that modulate HIV gene transcription and viral latency by using the GeCKO library. B. Scatter plot showing enrichment of sgRNA in activated or latent cells relative to control unsorted latent cells. Plot was generated by the CRISPR analyzer. Differences in enrichment were calculated as log2-normalized fold of change. The indicated colored sgRNAs were significantly enriched in our screen; of these the highest-ranking gene was ZNF304. This gene was detected across two independent screens and was thus chosen for further validation and mechanistic investigation. Dots near the diagonal represent sgRNA with no enrichment between latent or activate state of HIV.