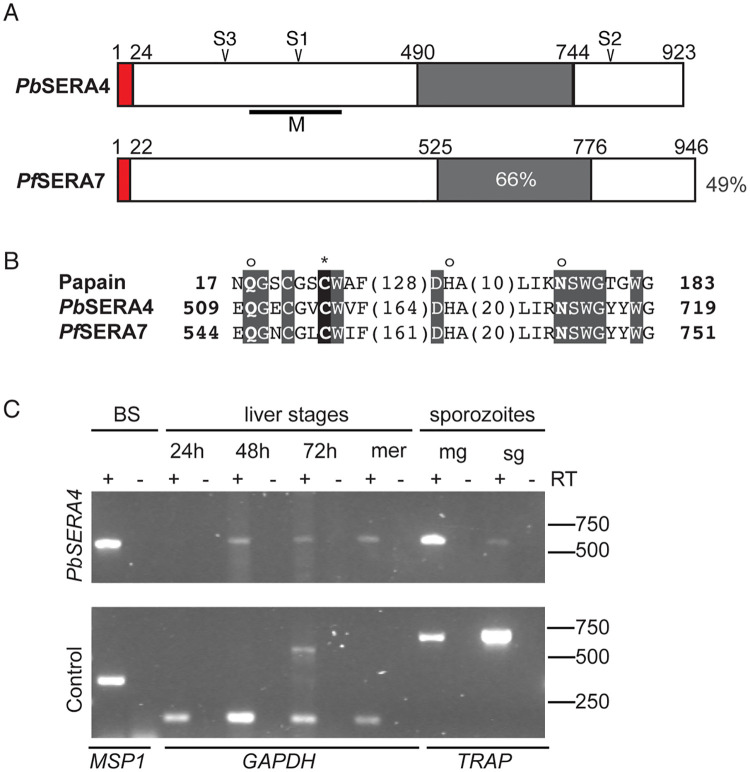
Fig 1. Plasmodium berghei SERA4 is a papain-like cysteine protease expressed across the Plasmodium life cycle.
(A) Schematic structure of P. berghei PbSERA4 (PBANKA_0304800) and its ortholog in P. falciparum PfSERA7 (PF3D7_0207400). The putative signal sequences and the central papain-like cysteine protease domains are boxed in red and grey, respectively. Putative cleavage sites S1, S2 and S3 are as indicated at amino acids 295 (IKGEDDLD), 806 (ISGQTDNQ) and 173 (LTAKIEED), respectively. Antibodies were generated against the portions of PbSERA4 indicated by the black line labeled M. Amino acid sequence identities between PbSERA4 and PfSERA7 are indicated as percent for both the overall sequence and the catalytic domain. (B) The catalytic residues of the papain family of cysteine proteases are conserved in PbSERA4 and PfSERA7. Strictly conserved amino acid residues are boxed in grey, and the putative active-site cysteine is boxed in black and marked with an asterisk. The catalytic histidine and flanking glutamine and asparagine residues, which form the oxyanion hole, are marked with an ‘o’. Papain is from Carica papaya. Bold numbers represent the amino acid positions within the protein before and after the catalytic domain; numbers in parenthesis represent the number of intervening amino acids at the indicated positions. (C) Expression profiling of PbSERA4 in P. berghei ANKA. RT-PCR analysis from mixed blood stages (BS), liver stages (at 24, 48, and 72 hours after infection), liver stage merozoites (mer), and midgut (mg) and salivary gland (sg) sporozoites. Control transcripts were merozoite surface protein 1 (MSP1), glyceraldehyde dehydrogenase (GAPDH) or thrombospondin-related anonymous protein (TRAP) for blood, liver, and mosquito stages, respectively. Products run at the expected sizes in comparison to these products amplified from P. berghei genomic DNA (S1A Fig).