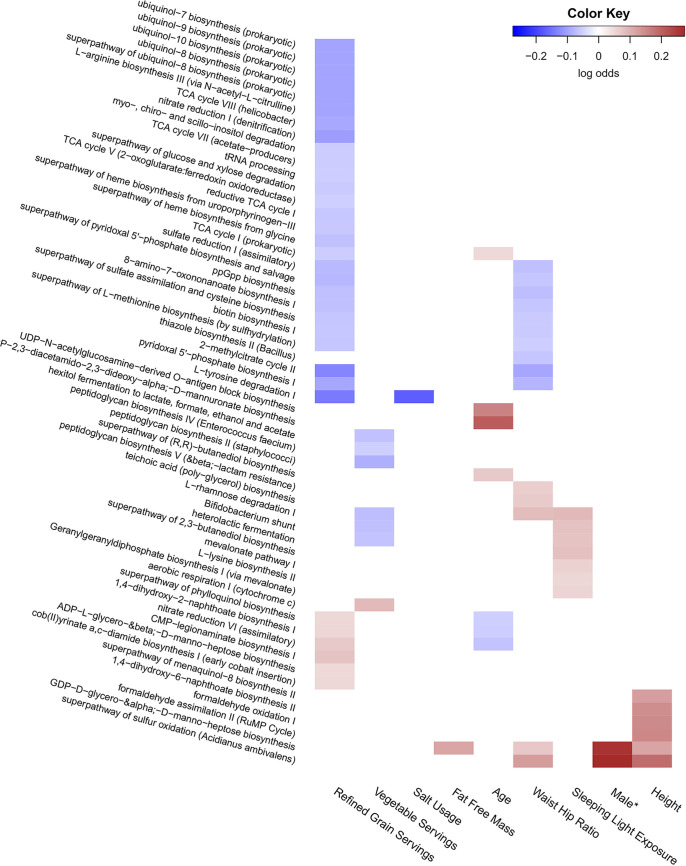
FIG 5.
Various predicted pathway abundances are associated with features significantly associated with overall microbiome composition. Pathway abundances were predicted from 16S rRNA gene sequencing data using PICRUSt2. Predicted pathway abundances meeting an FDR of <0.05 and an effect size of |0.05| log odds were considered significant associations using the Corncob R package. Each feature’s false discovery rate was corrected separately, and each was tested to control for differences in DNA extraction and differential variability within that feature. Ordinal variables were converted into a ranked scale for testing, and all features except for sex were scaled. The asterisk indicates that sex was treated as a categorical value; therefore, the magnitude is not directly comparable to other log odd ratios.