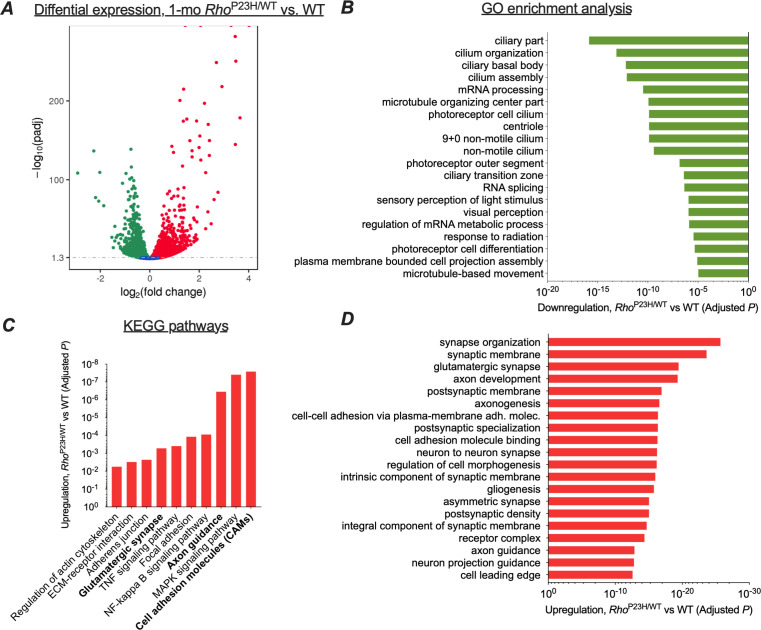
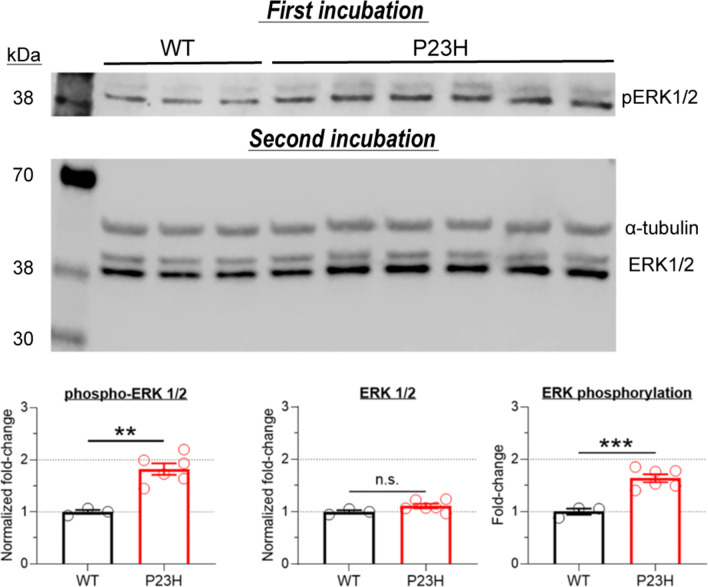
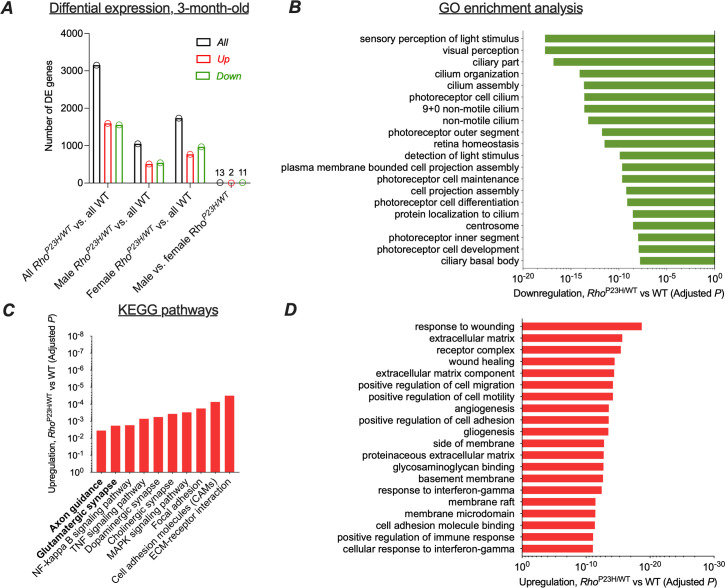
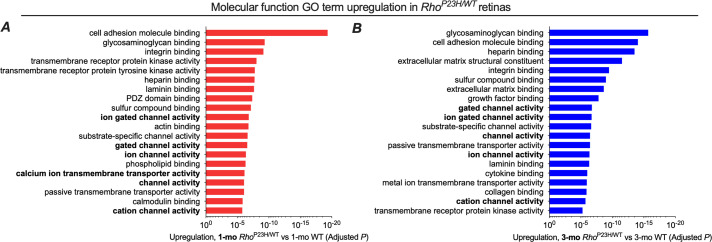
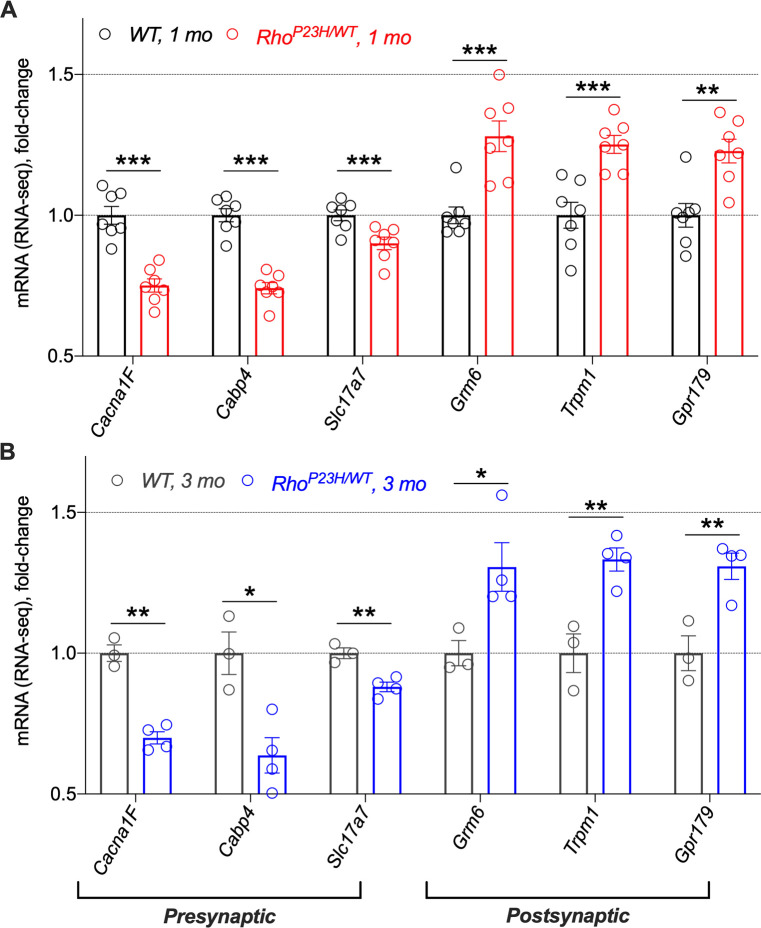
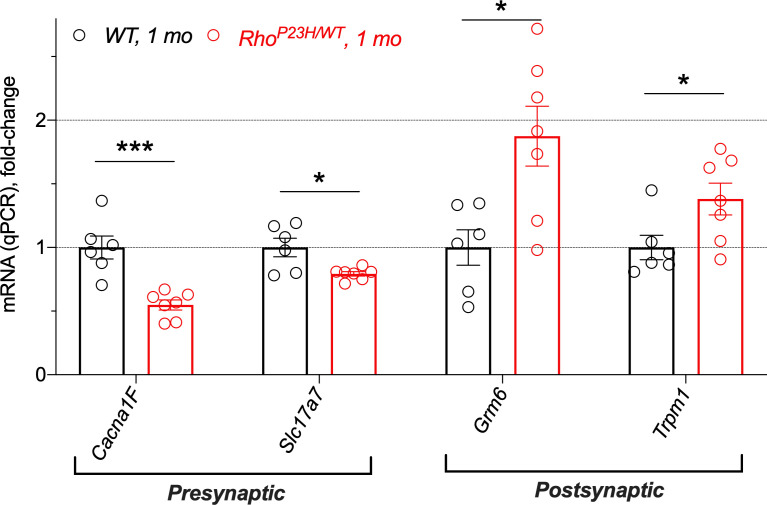
Figure 3. Transcriptomic analysis shows neural network adaptation to rod death in early stage retinitis pigmentosa.
RNA-sequencing was performed from whole retinal extracts at 1 month of age in P23H mice (n = 7) and WT (n = 7) littermates. (A) A total of 5404 transcripts showed differential expression (downregulation, green; upregulation, red) between P23H and WT retinas. (B) Gene ontology (GO) term analysis shows expected downregulation in photoreceptor-dominant gene clusters. (C) KEGG analysis shows significant enrichments in cell/neuron adhesion and growth pathways, glutamatergic synapse formation and several pathways associated with both cell stress and synaptic plasticity. (D) GO term analysis illustrates the most significant upregulation in synaptic and neural development and growth pathways in P23H retinas.