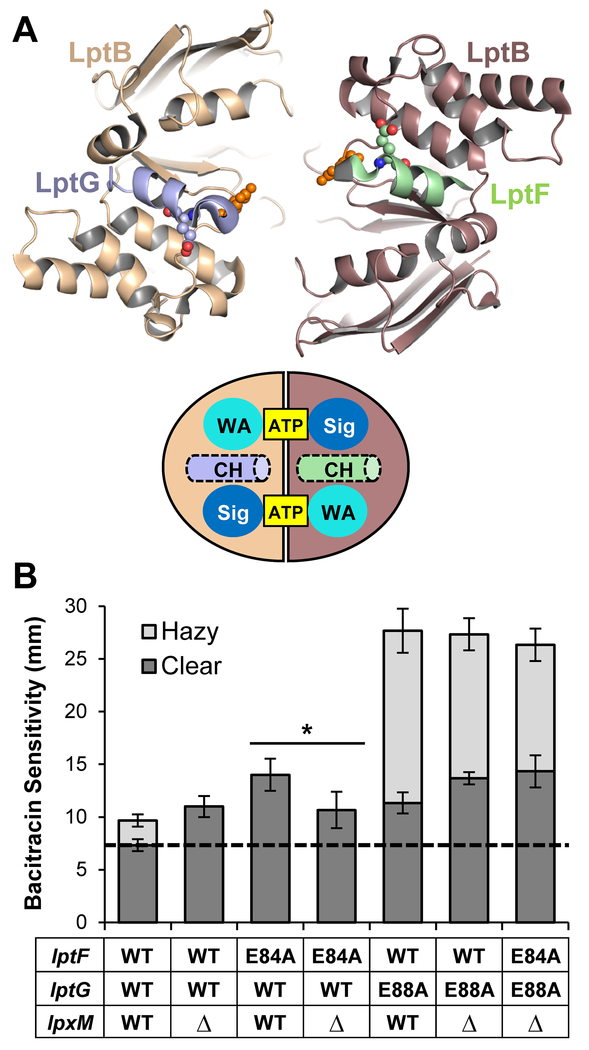
Figure 6: ΔlpxM suppress defects in the coupling helix of LptF.
A) Top: Top-down view from the membrane of the cartoon rendition of the Vibrio cholerae LptB2FGC structure (PDB 6MJP) cut away showing only the coupling helices of LptF (green) and LptG (light blue) and the LptB dimer. Residues E84 of LptF, E88 of LptG, and E86 of LptB are shown as spheres. Bottom: Drawing of the top-down view of the LptB dimer with the coupling helices (CH) pictured. ATP is bound between the signature (Sig) and Walker A (WA) motifs. B) ΔlpxM suppresses defects conferred by lptF(E84A) but not lptG(E88A). Plasmid-borne lptFG alleles were assessed for their ability to complement chromosomal ΔlptFG alleles in strains with chromosomal wild-type and deletion alleles of lpxM. OM permeability of viable mutants was assessed by sensitivity to bacitracin as measured by disc diffusion assay on LB plates. The haploid strain harboring the double glutamate mutant lptF(E84A) lptG(E88A) is unable to complement on LB, therefore it is not shown. The dashed line represents the diameter of the bacitracin disc. Data shown are the average of three independent experiments with error bars representing the standard deviation. NR2761 was used as the parent strain with wild-type lptFG and lpxM alleles. An unpaired two-tailed Student’s t-test was used to assess the significance of difference between lptF(E84A) and lptF(E84A) ΔlpxM. * indicates P < 0.05.