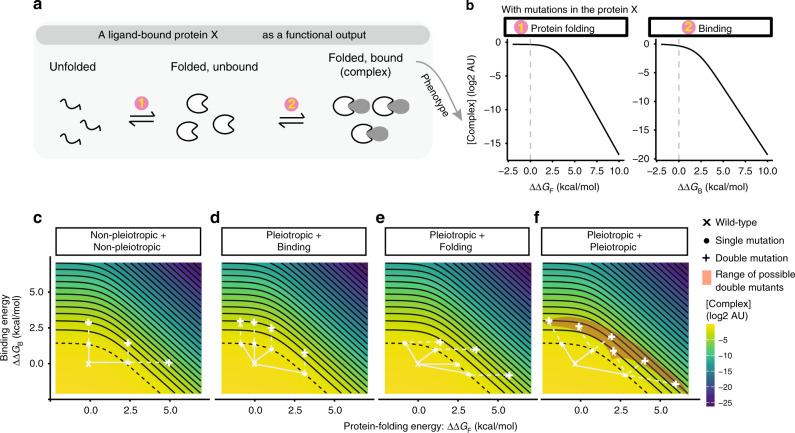
Fig. 6. Biophysical ambiguity in a protein–protein interaction system.
a Statistical thermodynamic model of a protein binding to a ligand. The protein X exists in three states: unfolded, folded, and folded and bound to the ligand. The partitioning of these molecules depends on the Gibbs free-energy differences between states. b Mutations result in additive changes in the free energy of protein folding and binding, altering the concentration of the protein–ligand complex. c–f Free-energy-phenotype landscapes for mutations that affect the free energy of folding (x-axis) and/or binding energy (y-axis). Phenotypic isochores are drawn with an interval of 1 in log(2) scale. A continuous range of free-energy changes can underlie an observed phenotype (dashed isochore). Combining two mutations with the same effect can result in a range of double mutant phenotypes (red shaded areas in (f)). Example double mutant outcomes are shown when neither (c), one (d, e) or both (f) mutations are pleiotropic. See also Supplementary Fig. 5. Source data are provided as a Source data file.