Figure 3 |. RALY and hnRNP-C are non-degraded substrates of Ad E1B55K/E4orf6-mediated ubiquitination.
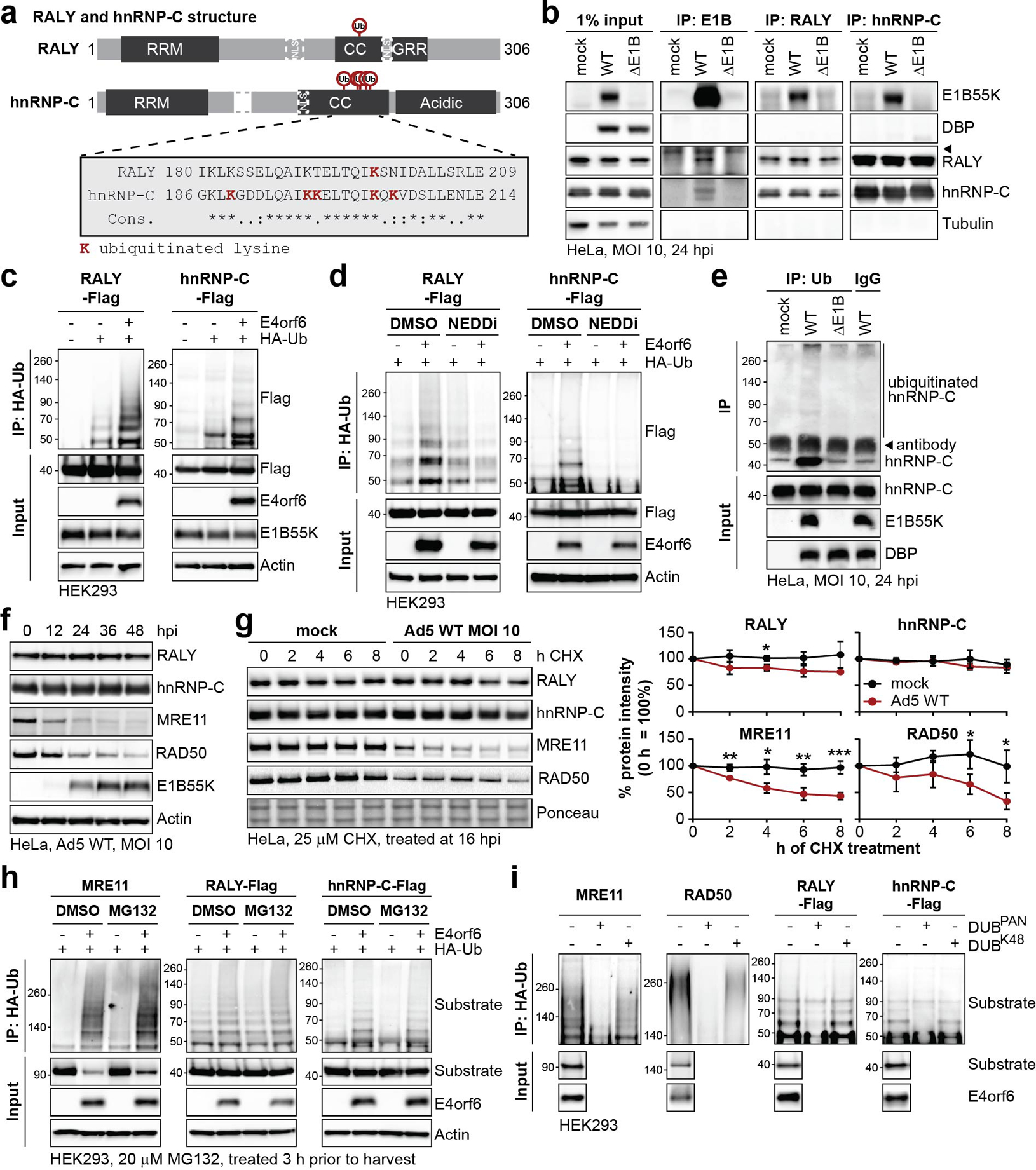
a, Domain structure of RALY and hnRNP-C. RRM=RNA recognition motif, NLS = nuclear localization sequence, CC=coiled-coil, GRR=glycine rich region. CC region shown below contains all detected lysine residues with increased ubiquitination upon E1B55K/E4orf6 expression highlighted in red. b, Immunoblot analysis of E1B55K, RALY, and hnRNP-C immunoprecipitations (IP) probing for pull-down of viral and cellular proteins during mock, WT and ΔE1B infection of HeLa cells at MOI of 10 for 24 h. DBP and Tubulin served as negative controls.◂ denotes antibody heavy chain. c, HEK293 cells transfected with indicated constructs for 24 h followed by denaturing IP with HA antibody and immunoblot analysis of RALY-Flag or hnRNP-C-Flag. d, HEK293 cells transfected with the indicated constructs for 24 h and treated with DMSO or NEDDi at 6 h prior to harvest, followed by denaturing IP with HA antibody and immunoblot analysis of RALY-Flag or hnRNP-C-Flag. e, Immunoblot of denaturing IP of hnRNP-C probing for ubiquitin during mock, WT and ΔE1B infections in HeLa cells at MOI of 10 for 24 h. ◂ indicates antibody heavy chain. f, Immunoblot analysis of protein levels over a time course of Ad5 WT infection (MOI=10) of HeLa cells. g, Immunoblot analysis and quantification of RALY, hnRNP-C, MRE11, and RAD50 over a time course of cycloheximide (CHX) treatment of mock or Ad5 WT infected HeLa cells. Shown is mean+SD. Statistical significance was calculated using an unpaired, two-tailed Student’s t-test, * p< 0.05, ** p <0.01, *** p <0.005. h, HEK293 cells transfected with the indicated constructs for 24 h and treated with DMSO or proteasome inhibitor MG132 at 3 h prior to harvest followed by denaturing IP with HA antibody and immunoblot analysis of MRE11, RALY-Flag, and hnRNP-C-Flag. i, HEK293 cells transfected with indicated constructs for 24 h followed by denaturing IP with HA antibody, treatment with the indicated deubiquitinating enzymes (DUBs) and immunoblot analysis of MRE11, RAD50, RALY-Flag, and hnRNP-C-Flag. All data are representative of three (b, e, g), four (d, h), or five (c) biologically independent experiments. Size markers in kDa are shown for ubiquitination immunoblots.
