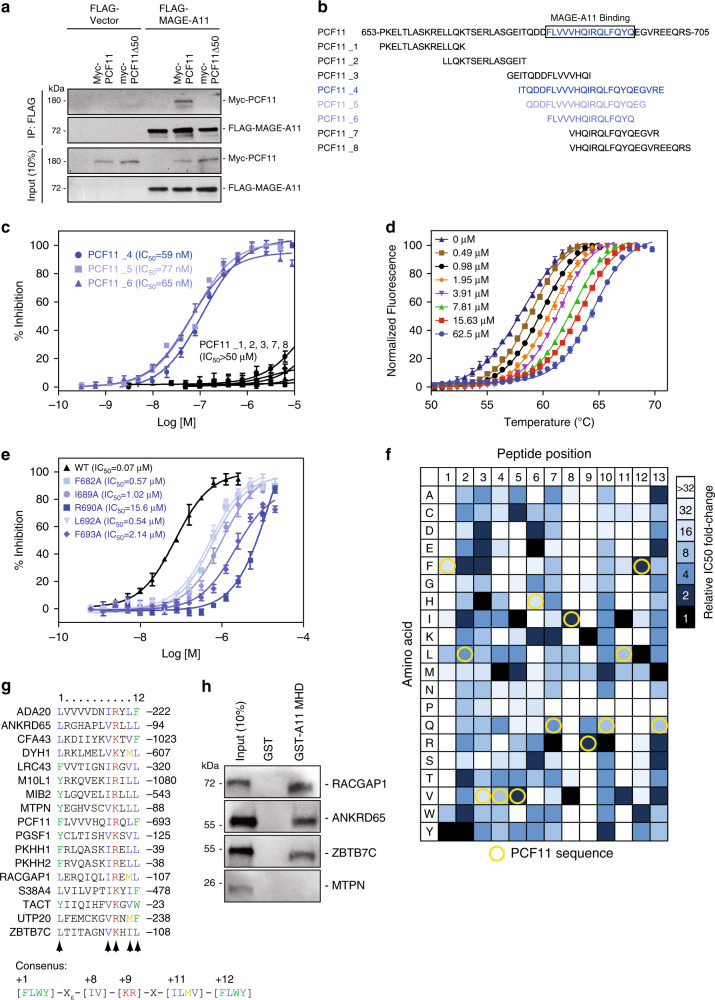
Fig. 1. Characterization of MAGE-A11 interaction with substrates.
a The MAGE-A11 recognition sequence in PCF11 is within amino acids 653-702. HEK293FT cells stably expressing FLAG-vector or FLAG-MAGE-A11 were transfected with PCF11 wild-type or PCF11 653-702 deletion construct for 48 h followed by IP with anti-FLAG, SDS-PAGE, and immunoblotting for anti-Myc. Source data are provided as a Source Data file. b, c Mapping of MAGE-A11-interacting motif in PCF11 by TR-FRET. Sequences of PCF11 degron motif and tested eight PCF11 peptides are shown (b). The binding activity of each peptide to GST-MAGE-A11 by TR-FRET is shown (c) (n = 2 biologically independent experiments with triplicates per peptide). Data are mean ± SD. d Validation of PCF11 peptide binding to MAGE-A11 by thermal stability assay with n = 3 biologically independent experiments per concentration. Thermal unfolding of MAGE-A11 is monitored with increasing concentrations of PCF11_6 peptide. Data are mean ± SD. e Alanine scanning mutagenesis of PCF11_6 peptide binding to GST-MAGE-A11 by TR-FRET (n = 2 biologically independent experiments with triplicates per peptide). Data are mean ± SD. f Relative TR-FRET affinity measurements of PCF11 degron peptide variants library (248 peptides) is shown. Coloring indicates relative binding affinity normalized at each position. Yellow circles indicates sequence of natural PCF11 degron peptide. g Comparison of 16 predicted MAGE-A11-binding motifs. h Recombinant GST-MAGE-A11 binds in vitro translated Myc-RACGAP1, ANKRD65, and ZBTB7C. Source data are provided as a Source Data file.