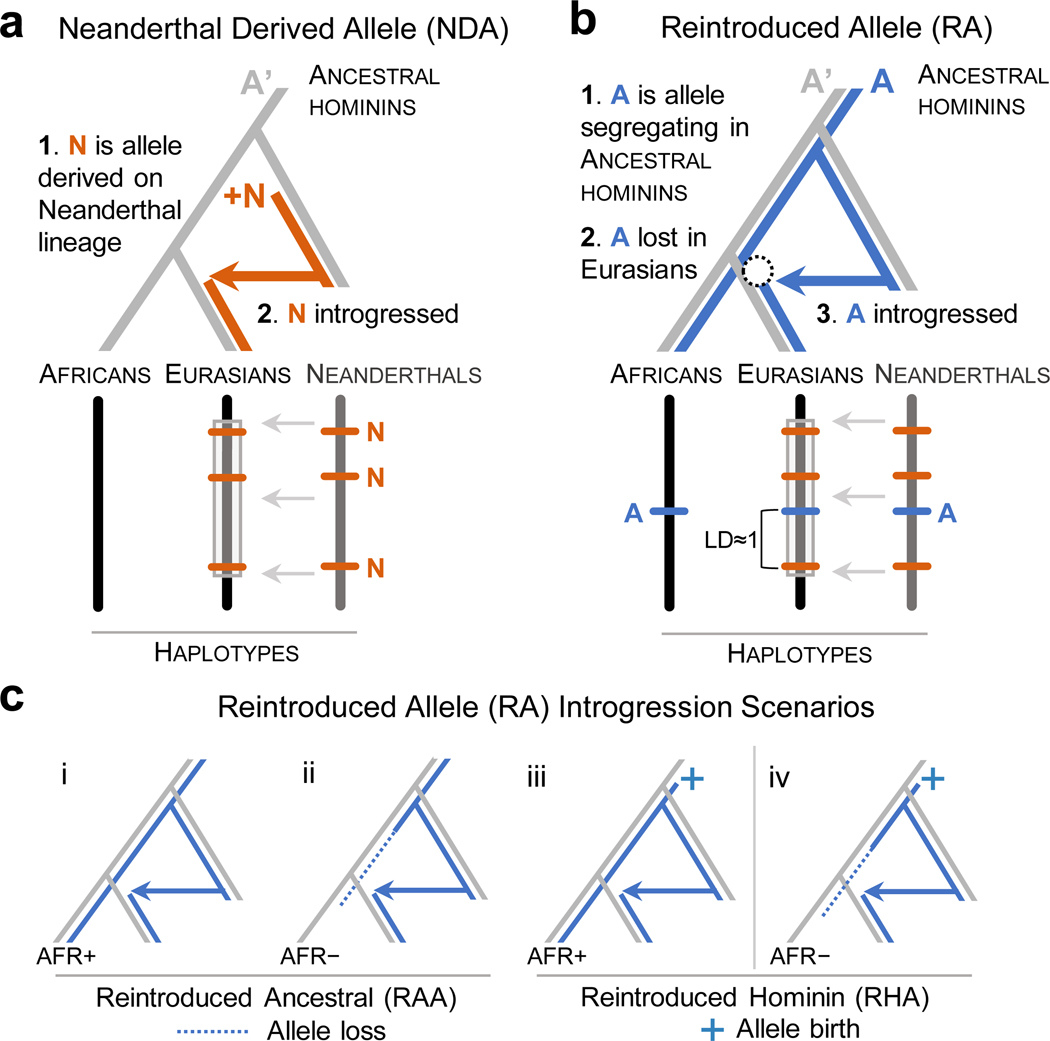
Figure 1. Schematic of the reintroduction of lost ancestral alleles by Neanderthal introgression.
(a) Evolutionary trajectory and resulting genomic signature of a Neanderthal derived allele N (orange) within an introgressed haplotype in modern Eurasians. A’ is the ancestral allele present in the population ancestral to anatomically modern humans (AMHs). (b) Evolutionary trajectory and resulting genomic signature of a reintroduced allele A (blue). To be an RA, A had to have been: (1) segregating (with A’) in the common ancestor of AMH and Neanderthals, (2) lost to the ancestors of modern Eurasians (dotted circle), and (3) reintroduced to modern Eurasians through Neanderthal admixture. RAs are characterized by their high linkage disequilibrium (LD≈1) with NDAs on introgressed haplotypes in modern Eurasians. (c) RAs can have different evolutionary histories. We distinguish several scenarios based on the status of the RA in the human-chimpanzee ancestor and in modern African populations. Reintroduced ancestral alleles (RAA, i and ii) are ancestral (i.e. they are the inferred ancestral allele of the human-chimpanzee ancestor) while reintroduced hominin alleles (RHA, iii and iv) appeared before the split AMH and Neanderthals, but may not have been present in the human-chimpanzee ancestor. Among RAAs, the ancestral allele may or may not be present in modern Africans (AFR+ vs. AFR–). However, the identification of RHAs requires that they are present in modern sub-Saharan African populations at reasonable frequencies (>1%). As a result, scenario iv cannot be inferred from modern genomic data alone and is not analyzed here.