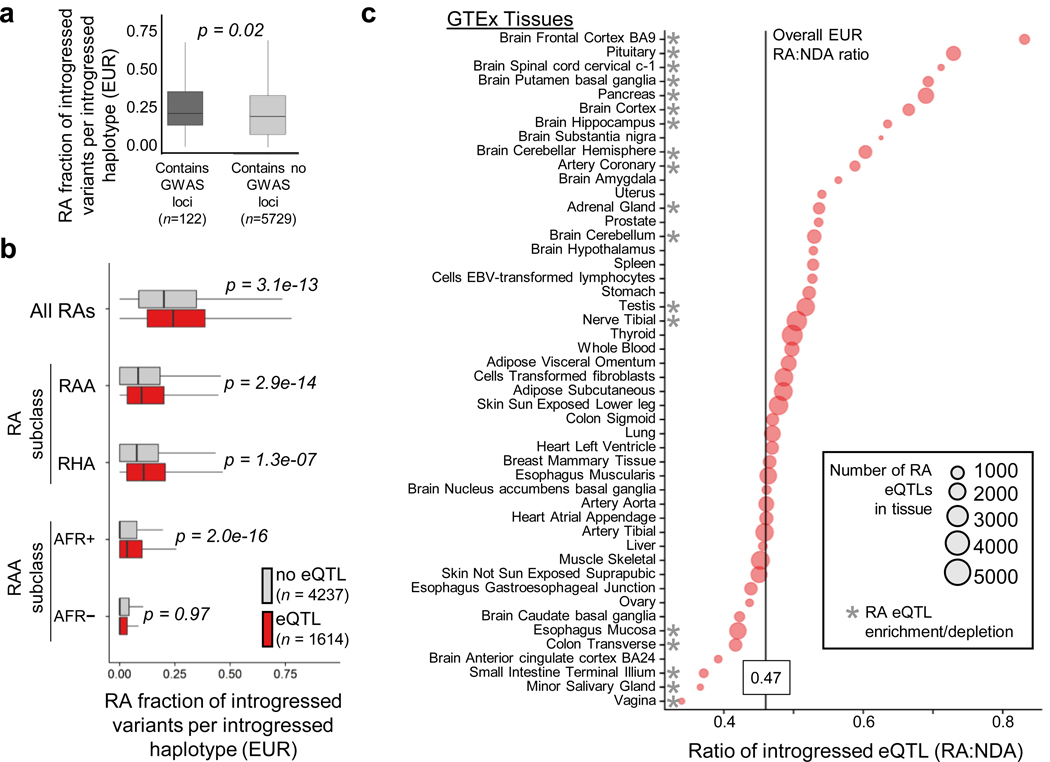
Figure 3. Reintroduced alleles are more prevalent than NDAs among introgressed haplotypes with GWAS hits and eQTL.
(a) The median fraction of RAs among all classified introgressed alleles (RAs plus NDAs) on introgressed haplotypes in Europeans (EUR) with at least one genome-wide significant (P < 1.0e−8) trait association reported in the GWAS Catalog versus introgressed haplotypes without trait associations (0.23 vs. 0.21 respectively, P = 0.02, Mann-Whitney U test). Introgressed haplotypes with GWAS hits have a higher RA fraction (and, symmetrically, lower NDA fraction) than those with no associations (median RA fraction of 0.23 vs. 0.21, P = 0.02). This enrichment varies across RA subclasses (Extended Data Fig. 7). (b) The fraction of RAs among introgressed alleles on introgressed haplotypes in EUR that contain GTEx eQTL (n=1,585) versus introgressed haplotypes without eQTL (n=4,237). Introgressed haplotypes with eQTL have a higher RA fraction than those with no eQTL (median RA fraction of 0.24 vs. 0.20, P = 3.0e−13, Mann-Whitney U test). This holds for all RA subclasses except RAAAFR–. (c) The RA:NDA ratio among introgressed eQTLs in each of 48 GTEx v7 tissues. Circles are scaled by the number of RA eQTLs in each tissue. Compared to the genome wide average RA:NDA ratio (0.47; vertical black line), 13 tissues have significantly more than the expected ratio of RA:NDA among eQTL and five tissues have fewer than the expected number (P < 0.01, hypergeometric test after Bonferroni correction). Brain tissues comprise eight of the 13 tissues with significant RA enrichment among eQTL.