Figure 1. Characterization of the BLOC1S5 deletion present in Patient 1.
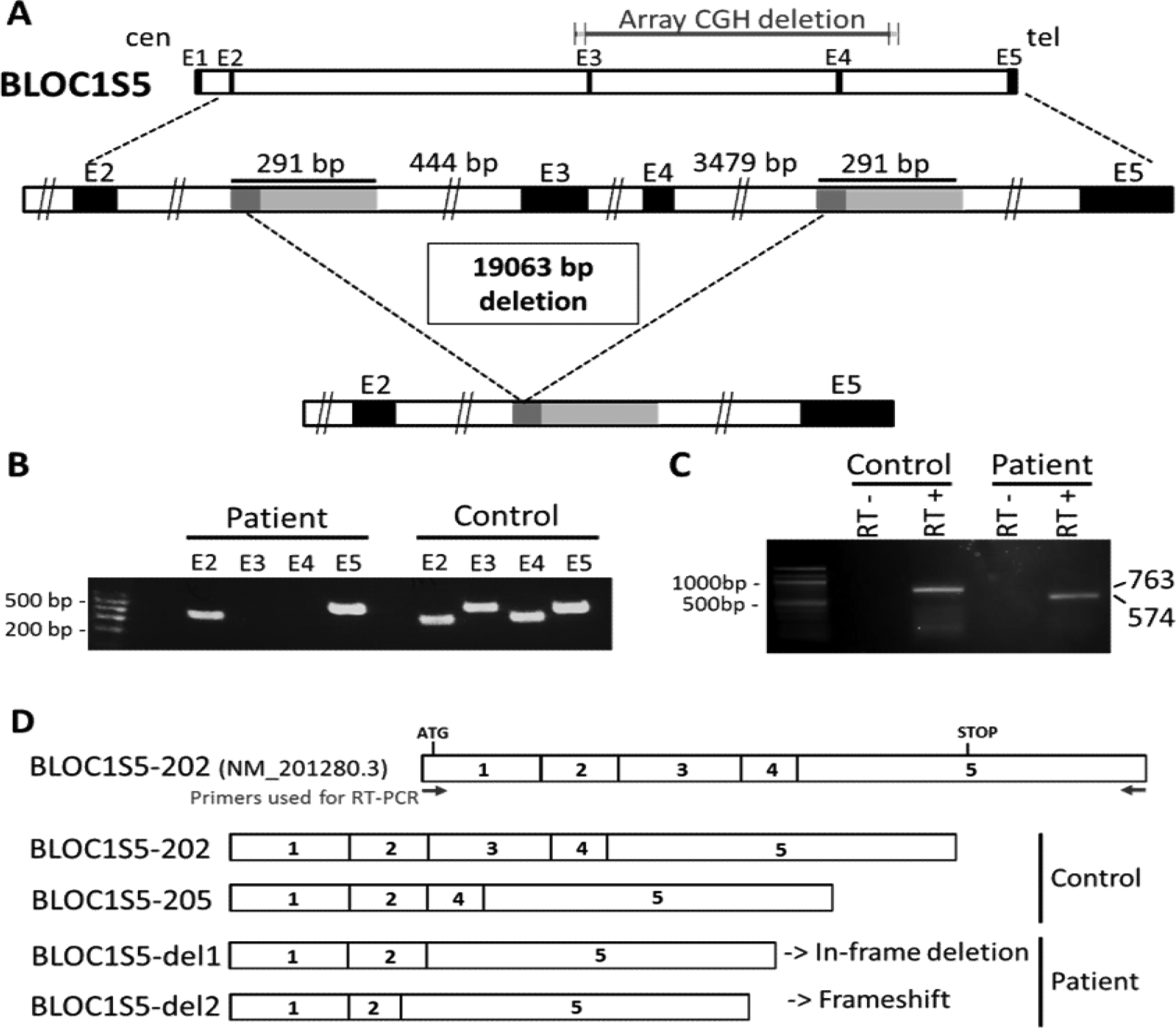
A) Schematic representation of the deletion. Top: genomic representation of the BLOC1S5 gene (approximatively to scale). The five exons are represented as black boxes. cen: centromeric side; tel: telomeric side. Middle: The region spanning exons 2 – 5 of the BLOC1S5 gene is expanded. The light gray boxes represent the 291 bp highly homologous segments (see Supplementary Data for details). The dark grey boxes represent the 57 bp perfectly homologous segment where non-allelic homologous recombination appears to have taken place in Patient 1. Bottom: Deleted allele resulting from non-allelic homologous recombination, showing that the region between the two 291 bp highly homologous segments is missing.
B) PCR amplification of exons 2, 3, 4 and 5 of BLOC1S5 in genomic DNA from a control individual and from Patient 1. A size marker (100 bp ladder) is shown on the left. Exons 3 and 4 are not amplified from the patient’s DNA.
C) Reverse-transcription PCR from leukocytes’ RNA of a control individual and Patient 1. RT-PCR primers where derived from BLOC1S5 exons 1 and 5, as shown in D). RT- : without reverse transcriptase. RT+ : with reverse transcriptase. Bands at 763 bp (corresponding to cDNA BLOC1S5–202) and at 574 bp (corresponding to cDNA BLOC1S5-del1) are observed in the control and the patient, respectively.
D) Schematic representation of the mRNA isoforms identified in the control individual and in Patient 1, as described in the main text. BLOC1S5–202 represents the theoretical wild-type isoform. Sizes of the complementary DNAs (cDNAs) corresponding to the different isoforms are BLOC1S5–202 (763bp) and BLOC1S5–205 (633bp), BLOC1S5-del1 (574bp), and BLOC1S5-del2 (542bp).
