Figure 7. Genetic Interaction between Rnd3 and Arhgap35 in Regulating CST Pathfinding.
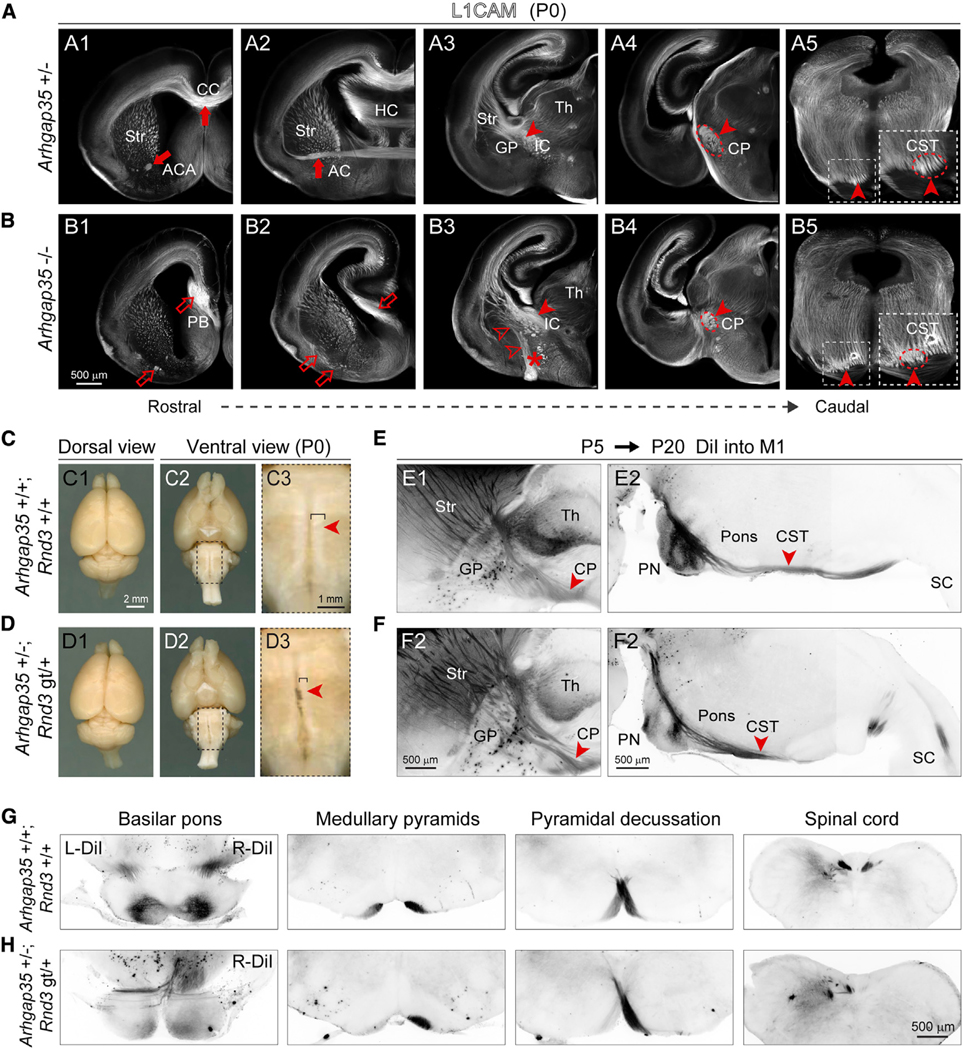
(A and B) Coronal sections from P0 Arhgap35+/− (A1–A5) and Arhgap35 −/− (B1-B5) brain stained for L1CAM reveal phenotypes, similar to Rnd3 gt/gt. The deficits are labeled with open arrows, arrowheads, and asterisks, whereas the normal trajectory is labeled with solid arrows and arrowheads. Scale bar: 500 μm.
(C and D) Gross morphology is maintained in Arhgap35 +/−; Rnd3 gt/+ brains, but the ventral brainstem shows thinner medullary pyramids (i.e., CST) as compared to wild-type controls. The closed arrowheads indicate the presence of the CST. Scale bars: 2 mm (C1 and C2, D1 and D2) and 1 mm (C3 and D3).
(E–H) DiI injection at P5 into primary motor cortex (M1) labeled the CST in CP and pons at P20 in Arhgap35 +/−; Rnd3 gt/+ brain depicting decussation defects in the sagittal (E1–F2) and in coronal sections (G and H) at medullary pyramids. The solid arrowheads indicate the CST decussation. SC, spinal cord. Scale bar: 500 μm.
See also Figures S9 and S10.
