Fig. 4. Mutational mechanisms of orthogene assimilation.
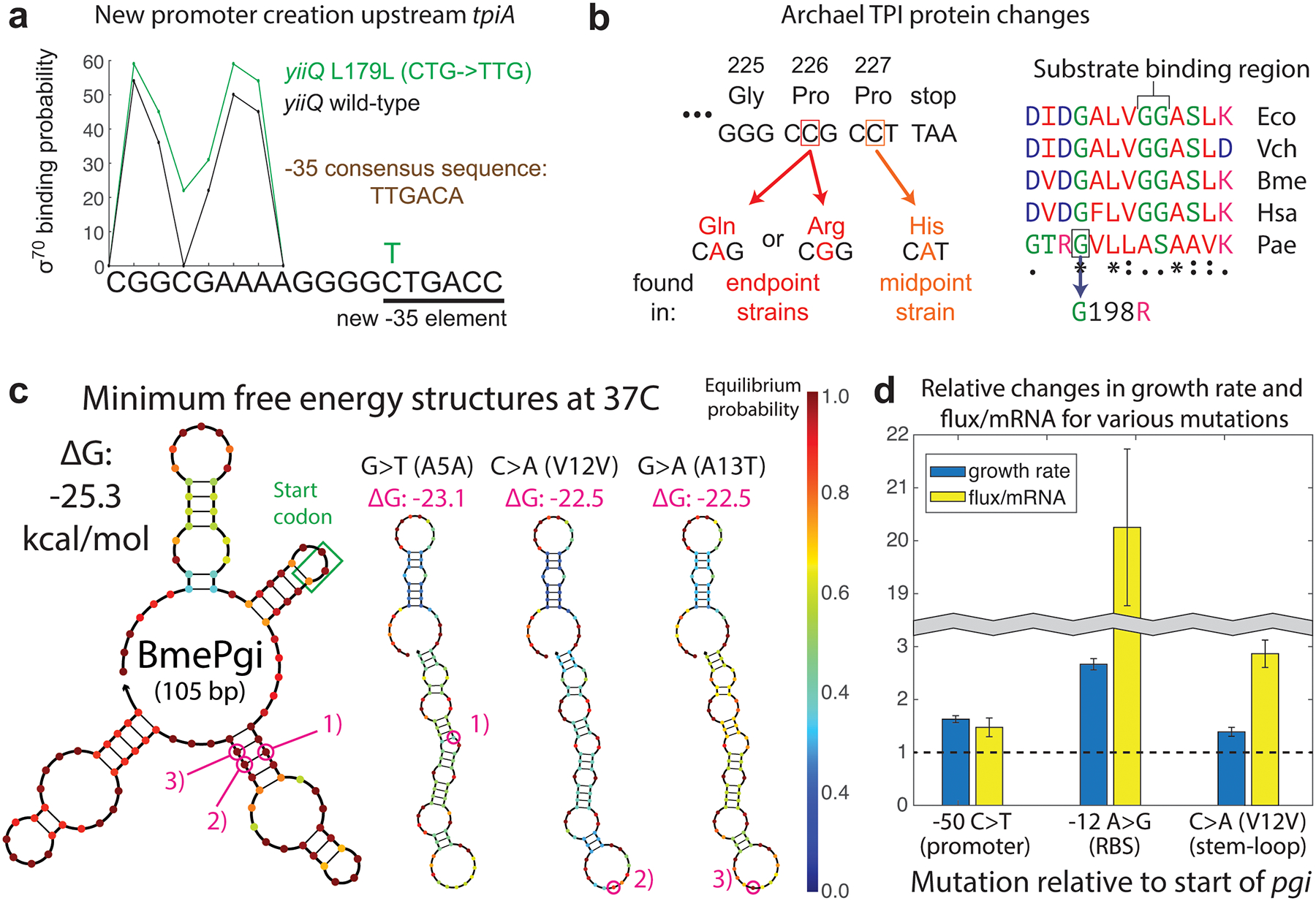
a, A promoter-creating synonymous SNP in the gene upstream of tpiA occurred independently more than 20 times. b, Missense SNPs in the archaeal TPI ameliorated polyproline ribosome stalling (left), and in one instance likely modified enzyme properties (right). c, N-terminal SNPs, frequently synonymous, targeted prohibitively strong stem-loops in mRNA secondary structure for destabilization. d, Various upregulating mutations had different impacts on the ratio of orthogene enzyme flux to mRNA level. Each pair of bars reflects growth, enzyme, and qPCR assays performed on two different strains – one lacking the mutation in question and one genetically identical except for the addition of the listed mutation. Error bars represent standard deviation from quadruplicate measurements.
