Figure 2. Identification and characterization of hyoscyamine dehydrogenase in A. belladonna.
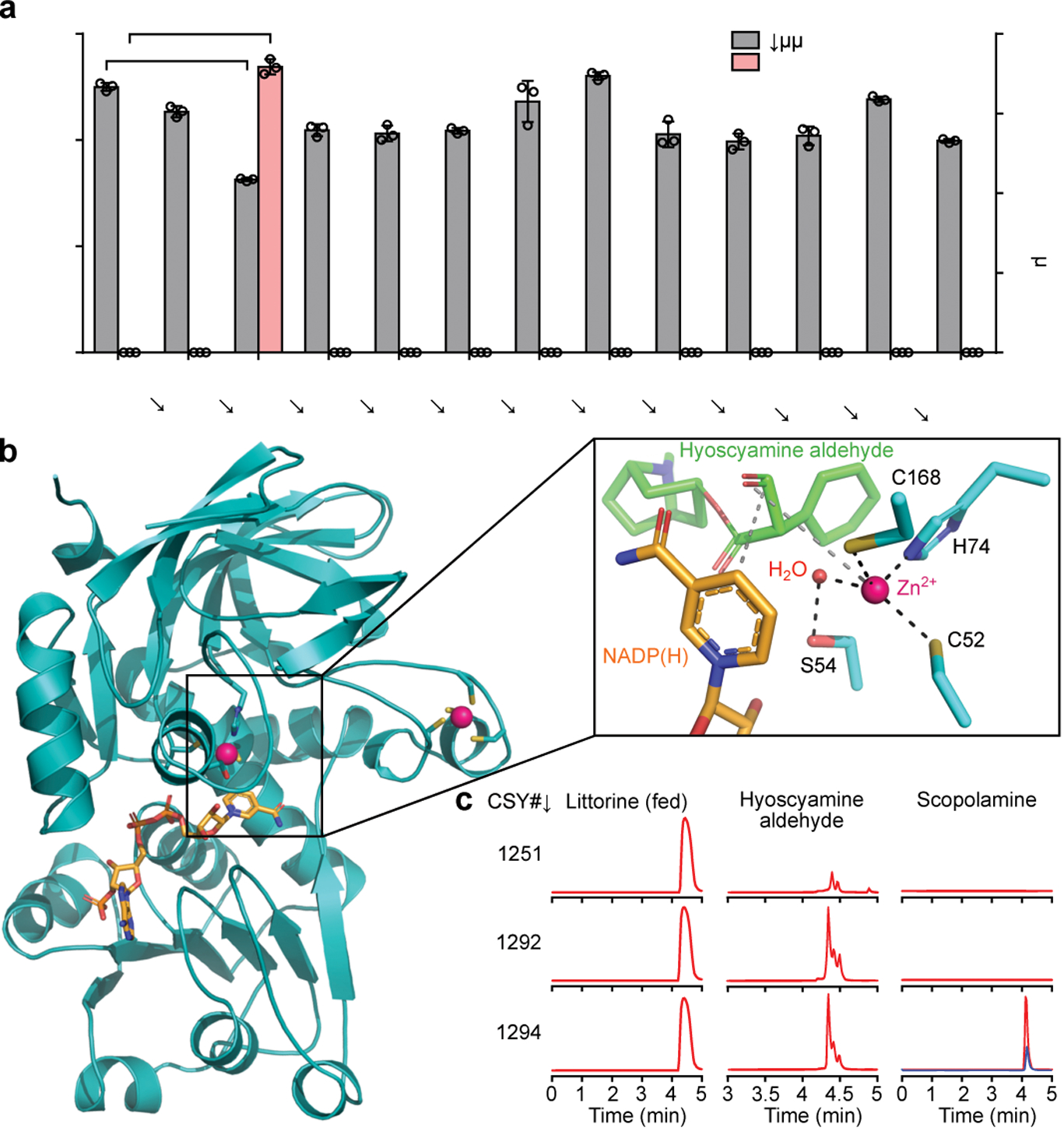
(a) Hyoscyamine aldehyde and scopolamine production in yeast engineered for expression of A. belladonna HDH candidates. Candidates or a negative control (BFP) were expressed from low-copy plasmids in CSY1292. Hyoscyamine aldehyde accumulation was compared using relative titers due to lack of an authentic chemical standard. Amino acid sequences are in Supplementary Table 1. Data represent the mean of n = 3 biologically independent samples (open circles), error bars show standard deviation. Student’s two-tailed t-test: *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance is shown relative to control. Exact P-values are in Supplementary Table 5. (b) Homology model of AbHDH. NADPH and Zn2+ are shown in orange and pink, respectively. Box: zoomed view of AbHDH active site with NADPH and docked hyoscyamine aldehyde. Dashed lines indicate interactions important for catalysis. (c) Multiple reaction monitoring (MRM) traces from culture media of yeast engineered for step-wise reconstitution of Module IV for conversion of littorine to scopolamine. Blue trace represents 125 nM (38 μg/L) scopolamine standard. Chromatogram traces are representative of three biological replicates. For a and c, strains were cultured for 72 h with 1 mM littorine prior to LC-MS/MS analysis of metabolites in culture supernatant.
