Extended Data Figure 4. Coexpression analysis, active site mutagenesis, and ortholog identification for AbHDH.
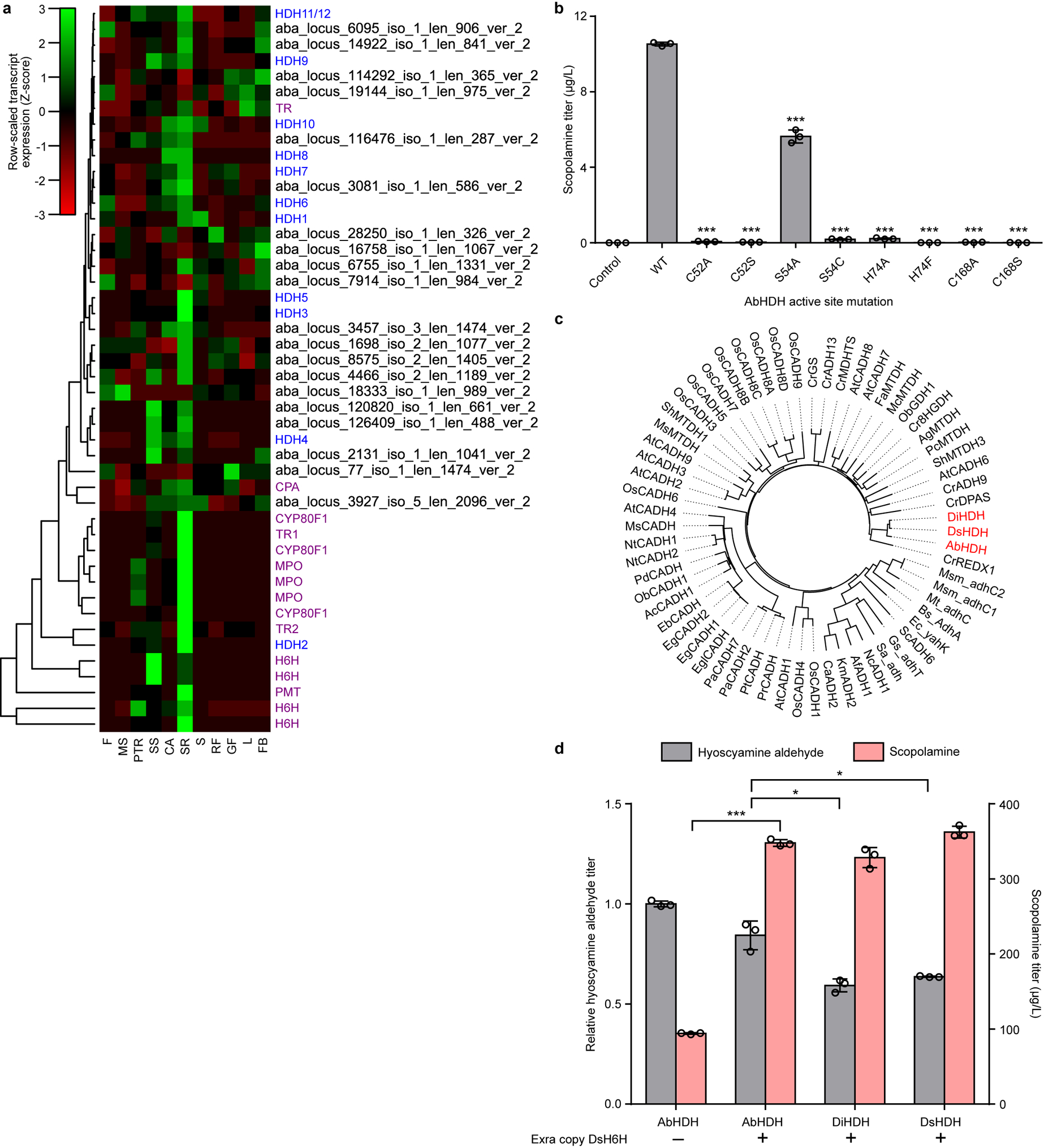
(a) Heatmap of tissue-specific expression profiles for hyoscyamine dehydrogenase (HDH) gene candidates identified from the A. belladonna transcriptome. Transcript expression is scaled by row using a normal distribution. Dendrogram indicates hierarchical clustering of candidates by tissue-specific expression profile. Color scheme for gene IDs: purple, known TA pathway genes; blue, putative HDH candidates with complete open reading frame sequences; black, putative HDH candidates with incomplete open reading frame sequences. Gene abbreviations (vertical axis): CPA, N-carbamoylputrescine amidase. Tissue abbreviations (horizontal axis): F, flower; MS, mature seed; PTR, primary tap root; SS, sterile seedling; CA, callus; SR, secondary root; S, stem; RF, ripe fruit; GF, green fruit; L, leaf; FB, flower bud. (b) Wild-type (WT) AbHDH, active site mutants, or a negative control (BFP) were expressed from low-copy plasmids in CSY1292. Transformed strains were cultured in selective media with 1 mM littorine at 25 °C for 72 h prior to quantification of scopolamine production by LC-MS/MS analysis of culture supernatant. Data indicate the mean of n = 3 biologically independent samples (open circles) and error bars show standard deviation. Student’s two-tailed t-test: *P < 0.05, **P < 0.01, ***P < 0.001; statistical significance is shown relative to wild-type. Exact P-values: C52A, 1.68 × 10−5; C52S, 2.06 × 10−5; S54A, 8.19 × 10−4; S54C, 5.27 × 10−6; H74A, 3.40 × 10−6; H74F, 2.78 × 10−5; C168A, 1.75 × 10−5; C168S, 2.39 × 10−5. (c) Phylogenetic tree of the three identified HDH orthologs (AbHDH, DiHDH, DsHDH) together with closest protein hits in the UniProt/SwissProt database. Phylogenetic relationships were derived via bootstrap neighbor-joining with n = 1000 trials in ClustalX2 and the resulting tree was visualized with FigTree software. Abbreviations: ADH, alcohol dehydrogenase; CADH, cinnamyl alcohol dehydrogenase; MTDH, mannitol dehydrogenase; DPAS, dehydroprecondylocarpine acetate synthase; 8HGDH, 8-hydroxygeraniol dehydrogenase; GDH, geraniol dehydrogenase; GS, geissoschizine synthase; REDX, unspecified redox protein. (d) HDH orthologs (AbHDH, DiHDH, DsHDH) were co-expressed with either a BFP negative control (‘–’) or an additional copy of DsH6H (‘+’) from low-copy plasmids in CSY1292. Transformed cells were cultured in selective media supplemented with 1 mM littorine for 72 h prior to LC-MS/MS analysis of culture supernatant. Data represent the mean of n = 3 biologically independent samples (open circles) and error bars show standard deviation. Student’s two-tailed t-test: *P < 0.05, **P < 0.01, ***P < 0.001. Exact P-values: AbHDH + DsH6H vs. AbHDH only, scopolamine, 4.68 × 10−5; DiHDH vs. AbHDH, hyoscyamine aldehyde, 0.0141; DsHDH vs. AbHDH, hyoscyamine aldehyde, 0.0372.
