Abstract
Unnecessary antibiotic regimens in the intensive care unit (ICU) are associated with adverse patient outcomes and antimicrobial resistance. Bacterial infections (BI) are both common and deadly in ICUs, and as a result, patients with a suspected BI are routinely started on broad-spectrum antibiotics prior to having confirmatory microbiologic culture results or when an occult BI is suspected, a practice known as empiric antibiotic therapy (EAT). However, EAT guidelines lack consensus and existing methods to quantify patient-level BI risk rely largely on clinical judgement and inaccurate biomarkers or expensive diagnostic tests. As a consequence, patients with low risk of BI often are continued on EAT, exposing them to unnecessary side effects. Augmenting current intuition-based practices with data-driven predictions of BI risk could help inform clinical decisions to shorten the duration of unnecessary EAT and improve patient outcomes. We propose a novel framework to identify ICU patients with low risk of BI as candidates for earlier EAT discontinuation. For this study, patients suspected of having a community-acquired BI were identified in the Medical Information Mart for Intensive Care III (MIMIC-III) dataset and categorized based on microbiologic culture results and EAT duration. Using structured longitudinal data collected up to 24, 48, and 72 hours after starting EAT, our best models identified patients at low risk of BI with AUROCs up to 0.8 and negative predictive values >93%. Overall, these results demonstrate the feasibility of forecasting BI risk in a critical care setting using patient features found in the electronic health record and call for more extensive research in this promising, yet relatively understudied, area.
Keywords: Critical Care, Prediction Models, Antibiotic Stewardship, Machine Learning, MIMIC, Electronic Health Records
Graphical Abstract
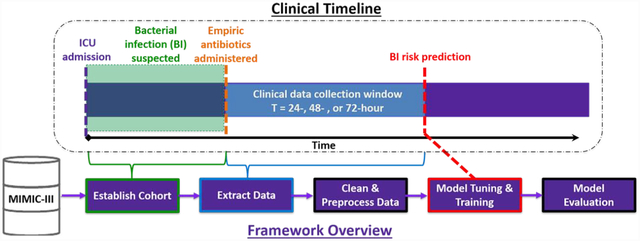
1. Introduction
Antibiotics can be life-saving for critically ill patients with bacterial infections (BIs), however, overuse or unnecessary administration can contribute to antimicrobial resistance (AMR) and antibiotic-associated morbidity.1–7 This is a critical issue, as patients with AMR infections suffer longer hospital stays, treatment complications, higher healthcare costs, and are more likely to die.8–11 Furthermore, antibiotics can cause harm through gut microbiome dysbiosis, mitochondrial toxicity, and immune cell dysfunction.1–7 Although clinicians have become more aware of the side effects of antibiotics, it is estimated that up to 50% of antibiotic prescriptions in acute care hospitals in the United States are still either inappropriate or unnecessary.12–17 Reducing both the amount and duration of unnecessary antibiotic treatments is a commonly proposed strategy to reduce the risk of antibiotic-related side effects.12–15,18 This is particularly relevant in the intensive care unit (ICU), where concerns for bacterial infections (BI) are high and prescribing antibiotics empirically—prior to having confirmatory bacterial culture results or when an occult BI is suspected—is a common practice.19,20
Approximately 30–50% of all ICU patients are diagnosed with a BI and their mortality rates can reach as high as 60% in severe infections.20–24 As a result, providers in the ICU often have a low threshold to start empiric antibiotic therapy (EAT) despite the ramifications of excessive antibiotic use for patients at low risk of BI. Unfortunately, there is no uniform consensus on the appropriate duration of EAT. As a result, clinicians must continually weigh the risks of failing to treat a serious BI against the risks of prescribing inappropriate antibiotic regimens. Moreover, physicians lack objective criteria to identify low BI risk in patients receiving EAT, and rely on clinical intuition and imprecise guidelines to balance EAT decisions.3,25–27 Strategies that shorten unnecessary antibiotic duration in ICU patients when BIs are no longer suspected offer a way to improve patient outcomes, and have been identified as a priority by the Society of Critical Care Medicine as part of their “less is more” campaign.28
Leveraging electronic health record (EHR) data with machine learning techniques presents an opportunity to accurately identify patients with low risk of BI. The widespread adoption of EHR systems offers investigators access to massive repositories of data generated through routine clinical care and provides opportunities to develop novel prediction algorithms to aid in clinical decision making.
The primary objective of this study was to develop a novel framework to identify ICU patients with a low risk of BI as candidates for earlier EAT discontinuation. The feasibility of this approach was investigated in patients suspected of having a BI by modeling data collected for up to 24-, 48- or 72-hours following the first dose of antibiotics. We compare prediction performance across different model types, data collection windows, and prediction thresholds. The developed algorithm could be used to identify patients at low risk of BI early in their hospitalization who may benefit from early discontinuation of EAT. Furthermore, our EHR-based phenotype of patients suspected of having a BI could be generalized to other datasets and used for additional analyses on antibiotic usage and BI in the ICU.
The detailed data dictionary, code, results been made available at: https://github.com/geickelb/mimiciii-antibiotics-opensource.
2. Materials & Methods
2.1. Dataset
A summary of our data extraction and analysis workflow is presented in Figure 1. The data used in this study was retrieved from the Medical Information Mart for Intensive Care III (MIMIC-III). The MIMIC-III database is an open and de-identified database comprised of health-related data from over 40,000 ICU patients who received care at Beth Israel Deaconess Medical Center between 2001 and 2012.29,30 MIMIC-III includes a variety of data such as administrative, clinical and physiological types, which are organized, formatted, processed and de-identified in accordance with the Health Insurance Portability and Accountability Act (HIPAA) guidelines.29,30
Figure 1.
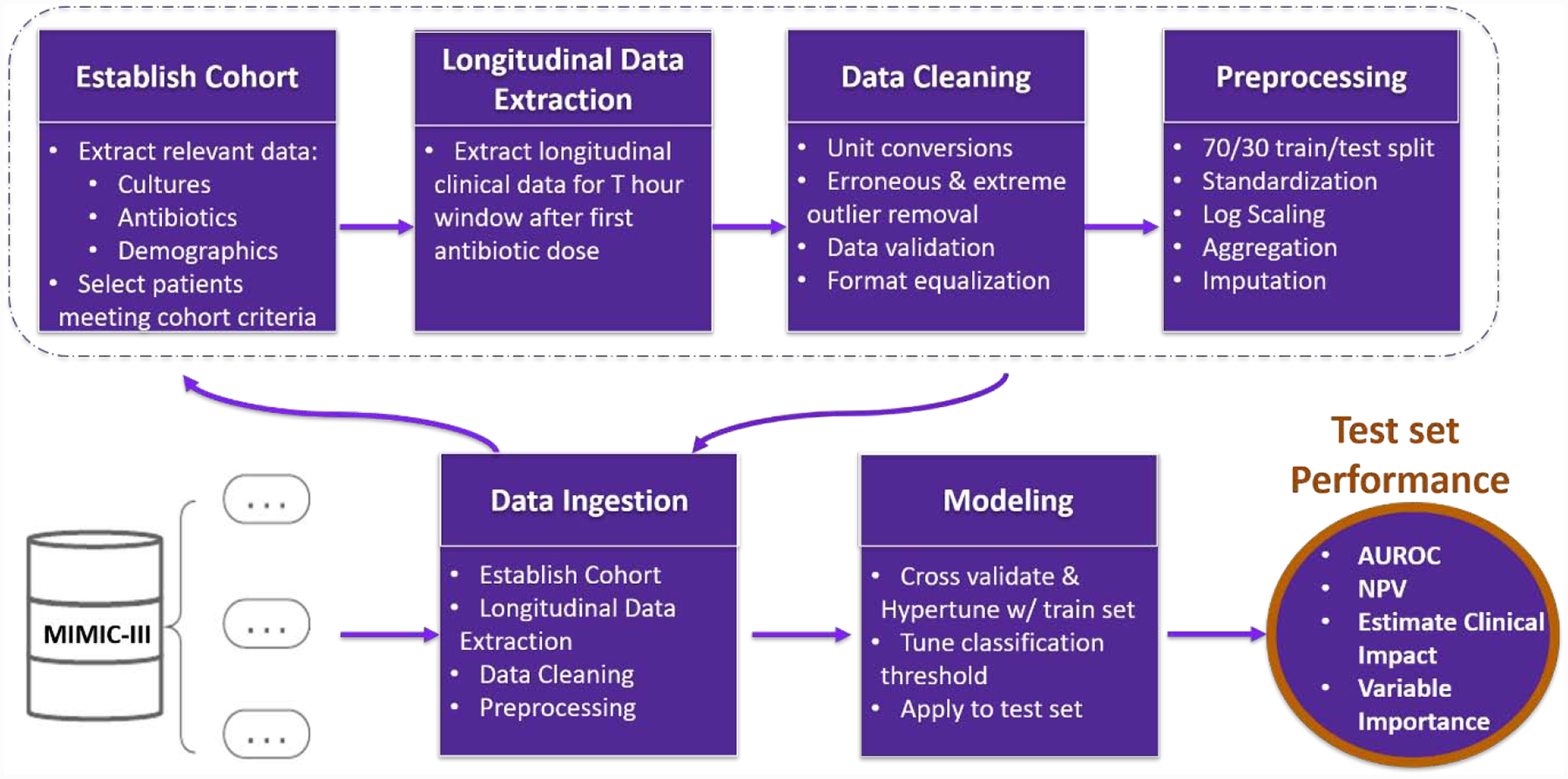
Data Ingestion and Analysis Framework Overview. Raw data is ingested from the MIMIC-III database. First a cohort of adult patients suspected of having SBI is established, and both longitudinal and categorical data is extracted over the T= 24-,48-, or 72-hour window following their first antibiotic dose that corresponds with an microbiologic culture. Next, data is cleaned, formatted, and preprocessed prior to modeling. The cohort is then filtered to patients with positive microbiologic culture and prolonged antibiotics, and microbiologic culture negative with short antibiotics. A 70/30 train/test set split is then applied. Scaling and standardization are performed on each set independently. Missing values were imputed using median values from the training set. Machine learning models are hypertuned on the training set and applied to the test set. Finally, classification thresholds are tuned, and model performance metrics are output.
2.2. Cohort
Adult patients who were suspected of having a BI upon admission to the ICU were eligible for our study. To match this phenotype, a patient must have: (1) received at least one dose of antibiotics within 96 hours following ICU admission and (2) had a microbiologic culture within 24 hours of their first antibiotic dose (Figure 2). Microbiologic cultures were defined as cultures obtained from any of the following: blood, joint, urine, cerebral spinal fluid (CSF), pleural cavity, peritoneum, or bronchoalveolar lavage. Patients with multiple ICU encounters that met study inclusion criteria were analyzed independently; however, each patient’s ICU encounters were assigned to the same train/test split (see Modeling).
Figure 2.
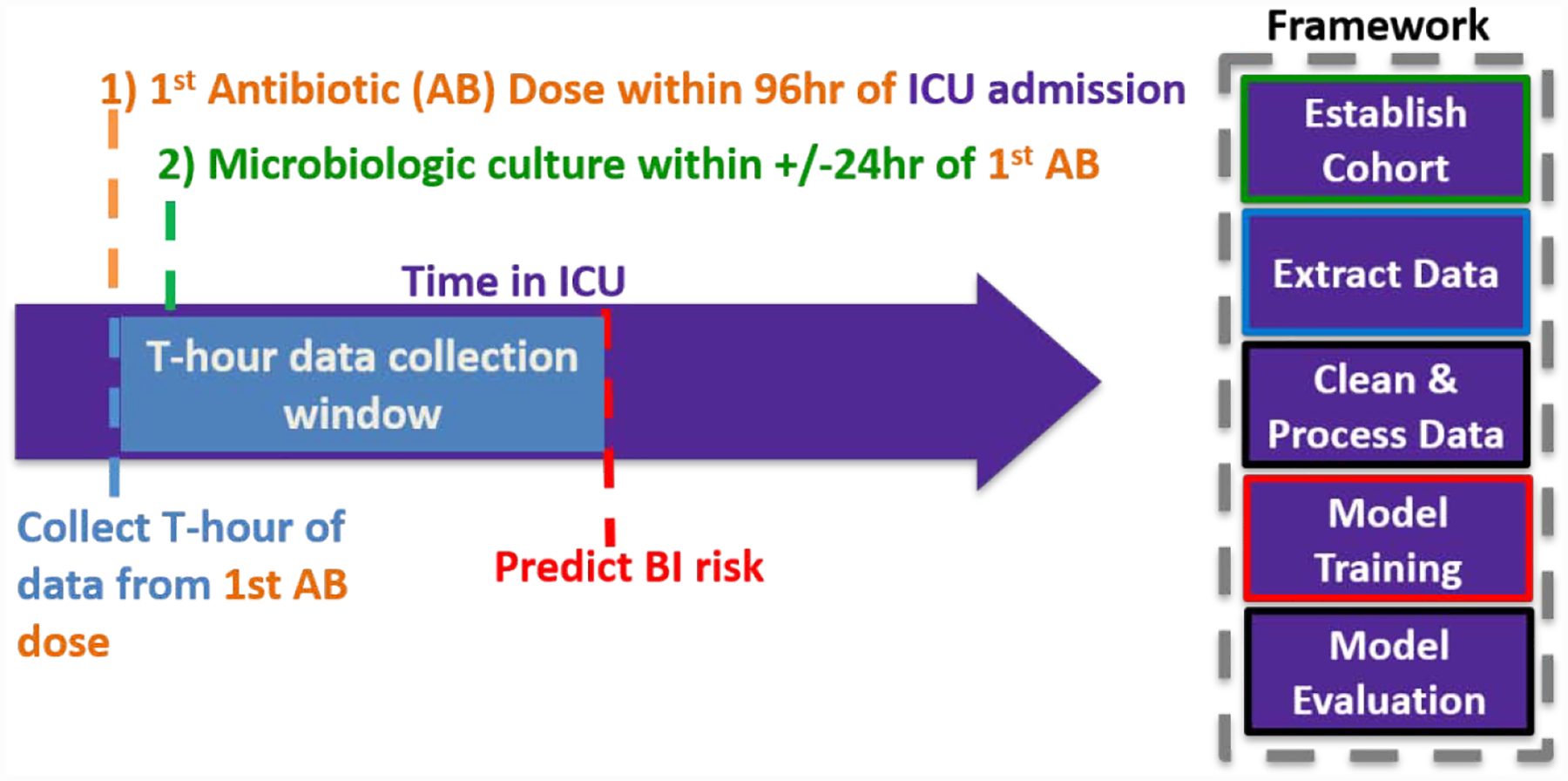
Phenotype Criteria for BI Suspicion at ICU Admission. A patient’s first Antibiotic (AB) dose (t0) needs to: (1) be administered within 96 hours following ICU admission and (2) have an microbiologic culture within 24 hours and (1) be administered within 96 hours following ICU admission. Clinical Data is collected for up to T= 24-, 48-, or 72-hours after first antibiotic dose.
Antibiotics prescriptions were recorded as the administration of any “antibacterial for systemic use” represented by Anatomical Therapeutic Chemical (ATC) code J01. ATC codes were obtained by first converting national drug codes (NDC) into RxNorm concept unique identifier (RXCUI) codes, and then into ATC codes. Regular expressions were used on prescription names to further filter out erroneous entries and those with missing NDC/RXCUI codes. We calculated the maximum length of cumulative antibiotic days following a microbiologic culture for each ICU encounter. Prescription information in the MIMIC-III database was stored with date level resolution. To accommodate this, the time of each patient’s first antibiotic dose (t0) meeting the phenotype criteria was set to 0:00:00.
Patients were allocated to one of three BI groups: serious BI, non-serious BI/no BI, and unknown BI status (Figure 3). Given the common occurrence of occult bacterial infections, a direct inference of BI status could not be made based off of microbiological culture results alone. Therefore, patient’s BI statuses were assigned based both on their microbiologic culture results (positive vs. negative) and duration of their antibiotic treatment (short [≤96 hours] vs. prolonged [>96 hours]). In this paradigm, patients with positive microbiologic culture and prolonged antibiotic treatment were considered to have serious BIs (prediction events), whereas those with negative cultures and short antibiotic treatment were considered to have no BIs (prediction non-events). Additionally, patients with short antibiotic treatment and positive microbiologic culture were considered non-serious BIs. Due to the possibility of occult infections, patients who received prolonged antibiotics despite having a negative microbiologic culture had less clear infection statuses, and were thus coded as unknown BI status. Conceptually, patients in that group could be further divided into those with an occult serious BI and those with either no BI or an occult non-serious BI. These patients were separated from the dataset prior to model training and testing, and were later used assess the clinical utility of the prediction model by testing its ability to identify patients at low-risk BI in that population.
Figure 3.
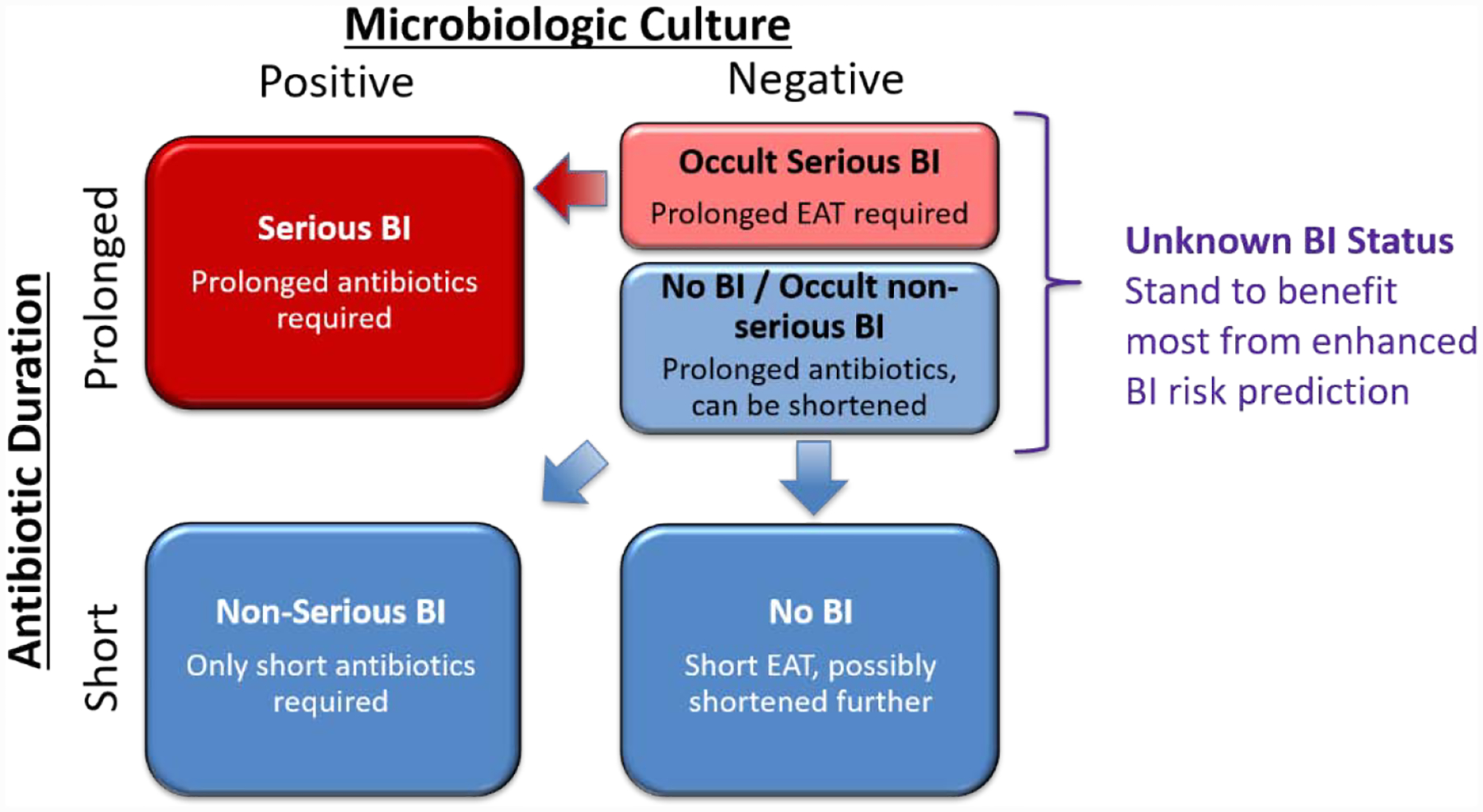
Classification of BI status and framing of the clinical prediction problem. Patient BI status can be classified into three groups based on duration of antibiotics and microbiological results: “Serious BI” are those with positive microbiological cultures receiving antibiotics for >96 hours and are the cases in model training. “Non-serious BI” and “No BI” patients are those with antibiotics ≤96 hours and are the controls in the model training. “Unknown BI status” are patient who received empiric antibiotic therapy [EAT] for >96 hours despite negative microbiological cultures, and are the group of patients most likely to benefit from correct BI risk prediction. The unknown BI status group may be conceptually divided into patients with “occult serious BI” who are likely more similar to the cases than to the controls, and patients with “no BI or occult non-serious BI” who are likely more similar to the controls than to the cases.
To control for Staphylococcus culture contamination, we required two consecutive Staphylococcus positive cultures to be considered microbiologic culture positive. Additionally, we coded patients that died within 24 hours of their last antibiotic dose as prolonged antibiotic treatment (n=1266). To accommodate for date-level resolution on prescription timings, we utilized a conservative 96-hour threshold for short vs prolonged antibiotic duration.
2.3. Data Extraction
We extracted static and longitudinal patient clinical data from the MIMIC-III database using open source code provided by the MIMIC-III team (Table 1). Longitudinal data was restricted to either the T= 24-, 48-, or 72-hour cutoff following the administration date of the first antibiotic dose (t0:t0+T) (Figure 2).
Table 1.
Extracted Data- Raw variables and units extracted from the corresponding table in the MIMIC-III database.
| MIMIC-III TABLE | Data Collected | Unit | % Missingness (T= 24–72 hour) |
|---|---|---|---|
| Diagnoses | ICD-9 codes (Elixhauser Comorbidity Index) | categorical | 0 – 0 |
| Admissions | Age | years | 0 – 0 |
| Ethnicity | categorical | 0 – 0 | |
| Gender | categorical | 0 – 0 | |
| ChartEvents | Blood pressure (systolic, diastolic) | mmHg | 0.2 – 0 |
| Glasgow Coma Scale | GCS score | 72.5 – 53.3 | |
| Glucose | mg/dL | 0.5 – 0.1 | |
| Heart rate | bpm | 0 – 0 | |
| Peripheral oxygen Saturation (SpO2) | % | 0 – 0 | |
| Temperature | deg. C | 1.6 – 0.2 | |
| Ventilation status | categorical | 1.3 – 0.9 | |
| Weight | kg | 8.4 – 8.4 | |
| InputEvents | Dobutamine | mcg/kg/min | 98.8 – 98.3 |
| Dopamine | mcg/kg/min | 94.9 – 94 | |
| Epinephrine | mcg/kg/min | 97.9 – 97.6 | |
| Norepinephrine | mcg/kg/min | 83.1 – 80.1 | |
| Phenylephrine | mcg/kg/min | 86.2 – 83.2 | |
| Renal replacement therapy | pos/neg | 0 – 0 | |
| Vasopressin | mcg/kg/min | 98 – 97 | |
| LabEvents | Bands | % | 87.3 – 82.6 |
| Serum bicarbonate | mEq/L | 2.1 – 0.3 | |
| Bilirubin | mg/dL | 60.1 – 47.8 | |
| Blood urea nitrogen (BUN) | mg/dL | 2 – 0.3 | |
| Serum chloride | mEq/L | 1.9 – 0.3 | |
| Serum creatinine | mg/dL | 2 – 0.3 | |
| Serum glucose | mg/dL | 0.5 – 0.1 | |
| Hemoglobin | g/dL | 2.6 – 0.3 | |
| International Normalized Ratio (INR) | ratio | 24.9 – 13.3 | |
| Serum lactate | mmol/L | 48.1 – 42.3 | |
| Urine leukocyte | pos/neg | 69.5 – 57.6 | |
| Urine nitrite | pos/neg | 69.5 – 57.6 | |
| Partial pressure of arterial oxygen (PaO2)/fraction of inspired oxygen (FiO2) ratio | ratio | 67.9 – 65.1 | |
| Partial thromboplastin time (PTT) | sec | 25.2 – 13.8 | |
| Partial pressure of arterial carbon dioxide (pCO2) | mmHg | 39.9 – 34 | |
| Serum pH | n/a | 41.9 – 36.7 | |
| Platelet count | K/uL | 2.6 – 0.3 | |
| Serum potassium | mEq/L | 1.6 – 0.3 | |
| White blood cell count | K/uL | 2.9 – 0.3 | |
| Serum calcium | mmol/L | 63.1 – 56.6 |
Threshold: threshold for manual review
SD denotes Standard Deviation
2.4. Cleaning & Pre-processing
The raw clinical data extracted for the purpose of this study were first cleaned and formatted to address data quality issues and then preprocessed to facilitate usability by selected machine learning models. The first cleaning step was to address desperate units of measurement by converting each variable into designated units (Table 1). Next, conservative thresholds were set to review erroneous values and data entry errors for removal based upon a combination of reference laboratory value limits, clinical knowledge, three sigma outlier criteria, and manual audit of a subset of free-text to confirm concordance. Finally, event and windowed continuous variables, such as administration of renal replacement therapy or mechanical ventilation were coded and discretized. The cleaned data were then converted into unit variances following as in (1), where is the median value of the patients with negative microbiologic culture and short duration EAT. Next, longitudinal and ordinal clinical variables spanning t0:t0+T were aggregated to produce single value(s) for each parameter using the operation that conferred the highest likelihood of infection (minimum, maximum or both). Lastly, categorical variables were encoded to dummy variables using the one-hot-encoding technique. The final dataset was represented by a 52-dimension feature vector.
| (1) |
2.5. Modeling
The patients with positive microbiological cultures and prolonged antibiotic duration (serious BI) and those with short antibiotic duration (no BI or non-serious BI) were split into a training and test set following a 70/30 split based upon unique ICU stay identifiers. Cohort splitting was performed on unique ICU stay identifiers where individual patients were sequestered to either the training or testing set to prevent testing set contamination. We chose to impute missing values with median values from the training set in order to facilitate implementation into a clinical setting. Empirical studies have suggested that including imputed values with high missingness can improve model clinical utility, so we chose to include imputed values with high missingness in our model (Table 1).31,32
The final dataset was modeled using a variety of machine learning algorithms, including Ridge regression33, Random Forests34, support vector classifier (SVC)35, extreme Gradient Boosted decision Tree (XG Boost)36, K-Nearest Neighbors, and Multilayer Perceptron (MLP). These models were chosen using a set of criteria that included each model’s relative interpretability, approach to handling nonlinearity, and ability to model categorical and continuous features. A soft voting classifier, or ensemble of all other models, was also used to test for significant performance gains or losses.
Class imbalance was addressed by classification threshold tuning and modeling specific class balancing parameters, such as bootstrapping and class weights, during hyperparameter tuning in order to simplify the modeling workflow. Modeling hyperparameters were tuned using 10-fold cross validation with a binary cross entropy loss function on the training set. The binary classification threshold was tuned in 10-fold cross validation to achieve a high sensitivity (sensitivity ≥ 0.9) and was averaged across all folds. This high sensitivity was chosen in order to reduce the number of false negatives and predict low BI risk with higher certainty. Threshold tuned model performances were assessed on the test set using area under the receiver operator curve (AUC), F1 score, negative predictive value (NPV), precision, and recall.
3. Results
3.1. Cohort
We identified a total of 19,633 ICU encounters (15,412 unique patients) in the MIMIC-III data that met inclusion criteria for our study. Within this set, we filtered our cohort down to 12,232 ICU encounters (10,290 unique patients) that had either prolonged antibiotics and positive microbiologic culture, or short antibiotics and negative microbiologic culture (Table 2). Table 3 summarizes the breakdown of these patients across the train/test splits. Additionally, 7,401 ICU encounters (6,520 unique patients) with unknown BI status (prolonged antibiotics and negative microbiologic culture) were set aside to test the prediction model’s ability to identify patients at low risk BI in that population.
Table 2.
Demographics- Distribution of cohort demographics.
| Variable | Mean/stdev. |
|---|---|
| Gender- N, % | |
| Female | 5709 (47%) |
| Male | 6523 (53%) |
| Age (yr) | 64.7 +\− 17.0 |
| Ethnicity- N, % | |
| African-American | 1385 (11%) |
| White | 8855 (72%) |
| Hispanic | 507 (4%) |
| Other | 1485 (12%) |
SD denotes Standard Deviation
Table 3.
Cohort Split- Breakdown of the train/test split against patient classes
| Microbiologic Culture | Antibiotic Durationa | BI Status Classification | Train No. (%) | Test No. (%) | Total No. (%) |
|---|---|---|---|---|---|
| Negative | Short | Positive | 5512 (65%) | 2355 (65%) | 7867 (65%) |
| Positive | Prolonged | Negative | 1693 (20%) | 745 (20%) | 2438 (20%) |
| Positive | Short | Negative | 1296 (15%) | 631 (15%) | 1927 (15%) |
| Negative | Prolonged | Unknown | N/A | N/A | 7401 (100%) |
Time on antibiotics, short (≤96 hours) vs. prolonged (>96 hours)
Table 4 summarizes the test set results for each threshold tuned model. The performance across the models for each T-hour test set showed little variation, where XGBoost and Random Forests slightly outperformed the other models in terms of AUC, F1 score, NPV, and precision. As the data window was increased from 24 to 72 hours, there were small increases in AUC across the best performing models for each time window. Figure 4 summarizes the ROC curve for all the T=24-hour models where all, except K-nearest neighbors, performed similarly. Additionally, when tested with the 72-hour test data, the 24-hour Random Forests model obtained an AUC of 0.787 (~0.013 increase). Similarly, the 72-hour Random Forests model produced an AUC of 0.765 (~0.028 decrease) when tested on the 24-hour data. These changes in AUC suggest that both the 24-hour and 72-hour models maintain similar model performances when making predictions on data collected over 48-hour longer and shorter collection windows, respectively.
Table 4.
Preliminary Model Results - Modeling parameters for each model on the test set using the high sensitivity threshold.
| Model | AUC | F1 | NPV | Precision | Recall | High Sensitivity Threshold |
|---|---|---|---|---|---|---|
| 72-hour Test set | ||||||
| Random Forests Classifier | 0.793 | 0.431 | 0.941 | 0.284 | 0.891 | 0.124 |
| XGBoost | 0.795 | 0.439 | 0.943 | 0.291 | 0.891 | 0.096 |
| MLP Classifier | 0.779 | 0.395 | 0.948 | 0.25 | 0.936 | 0.09 |
| Logistic Regression | 0.781 | 0.423 | 0.932 | 0.278 | 0.876 | 0.298 |
| SVC | 0.778 | 0.425 | 0.935 | 0.28 | 0.881 | 0.101 |
| K-NN | 0.734 | 0.357 | 0.936 | 0.219 | 0.963 | 0.04 |
| Voting Classifier | 0.793 | 0.429 | 0.946 | 0.281 | 0.905 | 0.147 |
| 48-hour Test set | ||||||
| Random Forests Classifier | 0.788 | 0.43 | 0.943 | 0.283 | 0.897 | 0.126 |
| XGBoost | 0.796 | 0.436 | 0.946 | 0.288 | 0.9 | 0.091 |
| MLP Classifier | 0.771 | 0.456 | 0.92 | 0.318 | 0.805 | 0.084 |
| Logistic Regression | 0.774 | 0.421 | 0.938 | 0.275 | 0.893 | 0.296 |
| SVC | 0.773 | 0.42 | 0.941 | 0.274 | 0.9 | 0.099 |
| K-NN | 0.733 | 0.393 | 0.922 | 0.252 | 0.887 | 0.044 |
| Voting Classifier | 0.788 | 0.436 | 0.939 | 0.29 | 0.881 | 0.147 |
| 24-hour Test set | ||||||
| Random Forests Classifier | 0.774 | 0.424 | 0.944 | 0.277 | 0.905 | 0.258 |
| XGBoost | 0.776 | 0.416 | 0.94 | 0.271 | 0.901 | 0.104 |
| MLP Classifier | 0.764 | 0.439 | 0.925 | 0.297 | 0.84 | 0.087 |
| Logistic Regression | 0.764 | 0.411 | 0.94 | 0.266 | 0.907 | 0.302 |
| SVC | 0.763 | 0.411 | 0.937 | 0.267 | 0.9 | 0.105 |
| K-NN | 0.714 | 0.382 | 0.922 | 0.243 | 0.903 | 0.044 |
| Voting Classifier | 0.776 | 0.421 | 0.939 | 0.275 | 0.895 | 0.177 |
Figure 4.
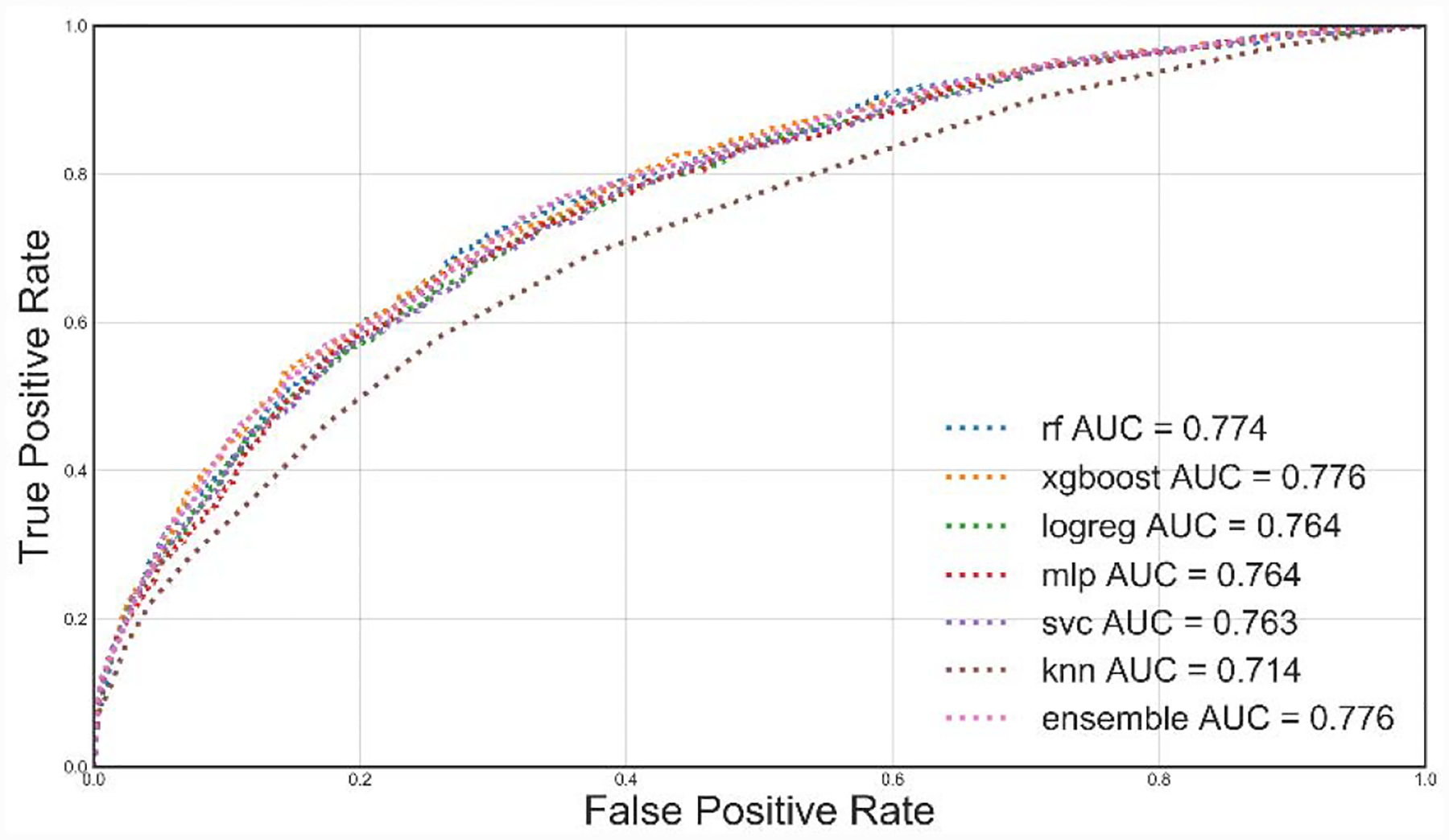
Receiver operating characteristic curves for all T=24-Hour models. We use different colors and line styles to differentiate models. AUC: Area under the curve.
Figure 5 displays how variable importance changed across the models. For this plot, a list of 20 variables was selected based on the top ten most important variables for the Random Forests, logistic regression, XGBoost, and SVC models. Figure 5 suggests that although the models perform similarly, each model prioritized predictors. This interpretation is reinforced by the results of the soft voting ensemble models, which performed comparably to the best performing model within each T-hour test set. This further suggests that the models are identifying the same or similar patients regardless of the underlying algorithm.
Figure 5.
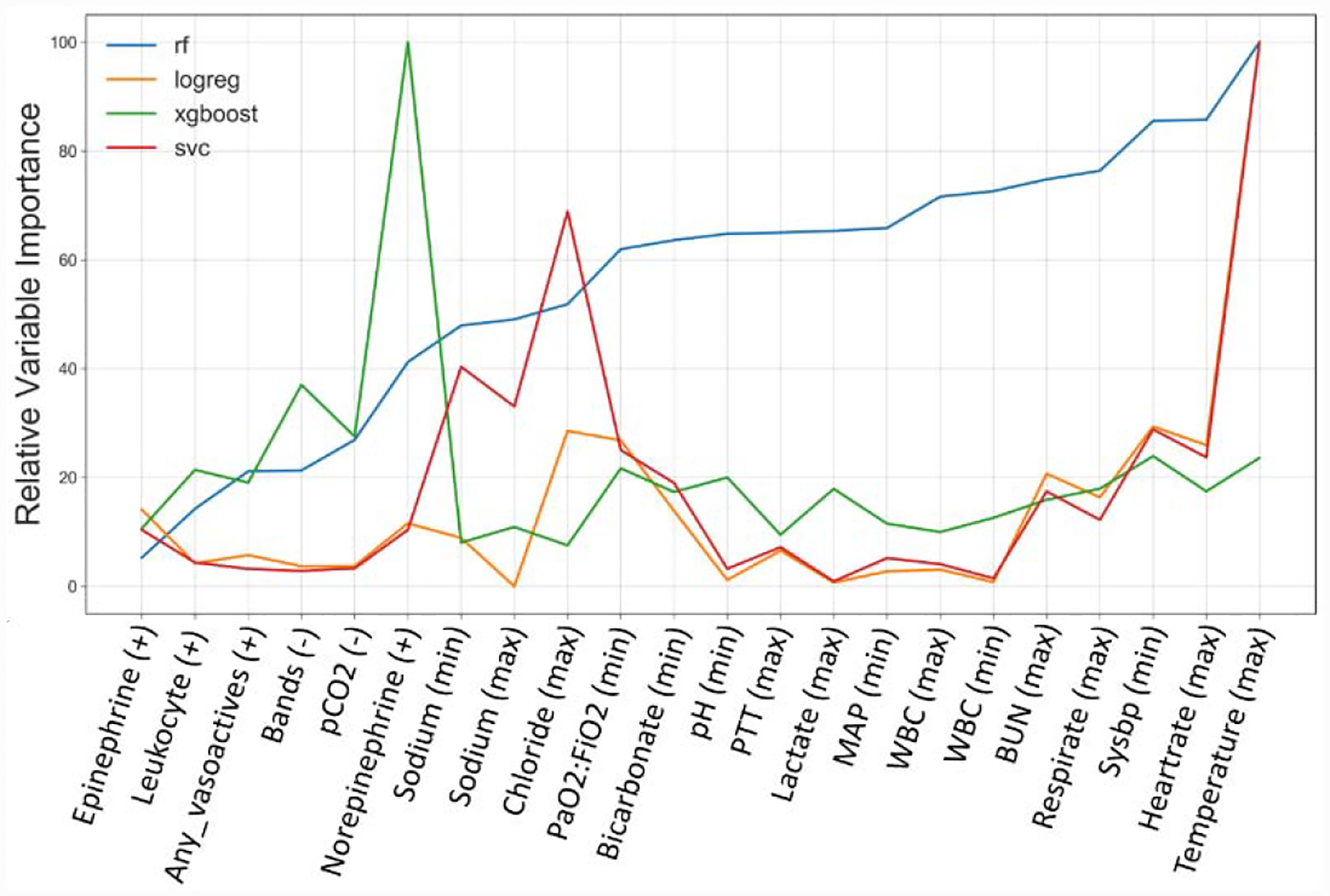
Stacked Relative Variable Importance Across Prediction Models. Variable importance for Random Forests and XGBoost were based on standardized Gini importance, while SVC and logistic regression used standardized coefficients. Variable importance values from all models were scaled relative to the value of the most important variable for all 20 values in the variable list. pCO2: carbon dioxide partial pressure; PaO2:FiO2: ratio of arterial oxygen partial pressure to fractional inspired oxygen; PTT: platelets; MAP: average arterial blood pressure over one cardiac cycle; WBC: white blood cell count; BUN: Blood urea nitrogen; sysBP: Systolic blood pressure
The T=24-hour Random Forests model was chosen for the subsequent analyses given that the T=24-hour timepoint provides more clinical utility, and thus the Random Forests model was the best performing model within this timepoint. Table 5. Confusion Matrix Statistics- Test set classification summary for the T=24-hour Random Forests model with a high sensitivity threshold. Table 5 summarizes the confusion matrix for this model with a high sensitivity classification threshold (0.26) in the test set. The model achieved an NPV of 0.944 in the test set; however, this figure is based on the 0.20 BI prevalence from the training and testing set. Figure 6 displays how the model NPV changes as a function of population BI prevalence and classification threshold. We found that as the BI prevalence changed from 0.5 to 0.1, the NPV of the T=24-hour random forests model changed from 0.82 to 0.98 when using a high sensitivity threshold, and 0.59 to 0.93 when using a 0.5 threshold. These results suggest that our model performance will be more robust to changes in prevalence when using the high sensitivity prediction threshold. The remaining patients falsely predicted as negatives by all of the T=24-hour models were investigated for observable patterns. These investigations suggested that the false negatives are a heterogeneous group with no reproducible patterns.
Table 5.
Confusion Matrix Statistics- Test set classification summary for the T=24-hour Random Forests model with a high sensitivity threshold.
| True Negatives (%) | False Positives (%) | False Negatives (%) | True Positives (%) | |
|---|---|---|---|---|
| High Sensitivity Threshold | 1208 (32.4%) | 1773 (47.5%) | 71 (1.9%) | 679 (18.2%) |
| 0.5 Probability Threshold | 2826 (75.7%) | 155 (4.2%) | 521 (14.0%) | 229 (6.1%) |
Figure 6.
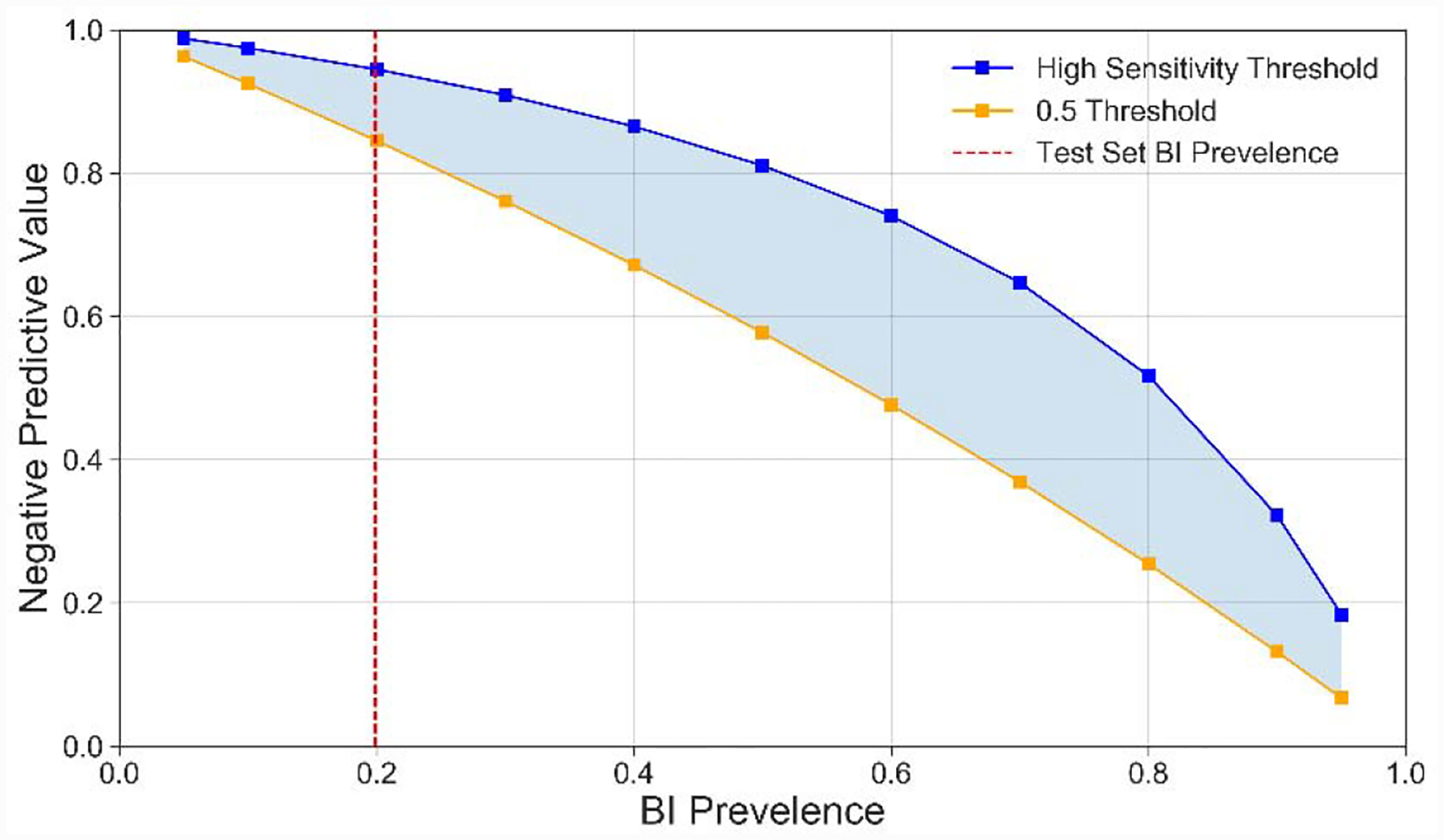
NPV across BI prevalence for T=24-hour Random Forests tuned and 0.5 prediction thresholds. NPV was simulated for a variety of BI prevalence values using the sensitivity and 1-specficity for the high sensitivity and 0.5 prediction thresholds from the test set.
Of the 1208 true negatives, 458 (37.9%) cases received antibiotics for 24 hours or less, while 750 (62.1%) received antibiotics greater than 24 hours. We estimated that 1,289 out of the 2,375(53.2%) total antibiotic days administered to patients in the true negative group could have been avoided if our model with a high sensitivity threshold were hypothetically used to stop EAT early.
3.2. Performance in patient set with unknown BI
Finally, the best performing T=24-hour Random Forests model was applied to the patient group with unknown BI status, which are those who stand to benefit the most from correct BI risk prediction. Using the high sensitivity and 0.5 probability thresholds, the model predicted 861 out of 7,401 (11.6%) and 5,525 out of 7,401 (74.7%) patients to be at low risk of BI, respectively (Table 6). Using the NPV from the test set with high sensitivity and 0.5 thresholds (NPV=94.5%, 84.3%) we estimated that approximately 48 (0.6%) and 860 (11.6%) of all unknown BI status patients would have been predicted to have a low BI risk but actually have had a BI (false negatives). By subtracting these estimated false negative patients from the total negative predictions, we estimated that the high sensitivity and 0.5 thresholds would have theoretically benefited 813 (11.0%) and 4,664 (63.0%) patients, respectively. We estimated that as a lower bound, our T=24-hour Random Forests model with a high sensitivity threshold could have reduced approximately 5,684 (9.5%) antibiotic days administered to this group, and as an upper bound with the 0.5 probability threshold could have reduced approximately 35,831 (60.0%) antibiotic days. A manual chart review and clinical assessment of 10 patient records with unknown BI status (5 predicted high BI risk, 5 predicted low BI risk) found that 8 out 10 model BI risk classifications matched the clinical reviewer’s assessment of BI risk, 2 out of 10 were probably correct but remained indeterminate, and 0 out of 10 were misclassified (Appendix 1).
Table 6.
Prolonged Antibiotic Negative Microbiologic Culture Predictions- Prediction distribution for the T=24-hour Random Forests model.
| High Sensitivity Threshold | 0.5 Threshold | |
|---|---|---|
| Predicted low BI risk | 861 (11.6%) | 5525 (74.7%) |
| Predicted high BI risk | 6540 (88.4%) | 1876 (25.3%) |
4. Discussion
In this study, we developed a novel framework to extract patient features from raw clinical data and identify patients at low risk of BI who, in theory, could benefit from earlier EAT discontinuation within 24 hours of initiation. Our main finding is that our models can predict patients with low risk of BI with good performance when applied to structured clinical data collected for T= 24-hours after the EAT initiation. We also found that increasing the data collection time and model complexity yielded only slight performance increases. Finally, our results suggest by that applying our T=24-hour Random Forests model with a high sensitivity threshold to the patient set with unknown BI status (prolonged antibiotics and negative microbiologic culture), we would be able to identify around 11.6% of patients as candidates for EAT removal with high confidence and could reduce total antibiotic days by approximately 9.5%.
Designing data-driven approaches to accurately stratify patients based on their BI risk has the potential to greatly improve antibiotic stewardship efforts. Antibiotic stewardship in the ICU can be viewed as a two-stage process. The first stage requires administering broad-spectrum antibiotics to maximize treatment of serious BI. In the second stage, physicians either stop EAT for patients at low risk of BI or narrow the spectrum of antibiotics once the infection is characterized.3 Many stewardship techniques focusing on the later stage hinge upon sensitive and specific identification and monitoring of BI risk. Bacterial cultures and inflammatory biomarkers are currently the most common methods of monitoring BI risk in the ICU, but are not necessarily optimal. Bacterial cultures, the current gold standard for diagnosing BI, may take days to result and are often unreliable in detecting all BIs.37 To address this, bacterial cultures are frequently supplemented with Gram staining, which provide additional information more immediately about a patient’s BI risk. However, Gram staining suffers from high variability and low reliability that results from individual differences in slide preparation and interpretation.38–40 Assays based on inflammatory biomarkers, such as C-reactive protein and procalcitonin, have improved sensitivity and specificity for detecting community-acquired infections, but have high rates of false-positives and -negatives for hospital-acquired infections.3,41–43 Newer rapid multiplex diagnostics for infectious organisms have also been introduced; however, these are still being tested for efficacy, costly, and not yet widely available.44 Designing better methods to identify patients with low risk for BI is critical to shorten the duration of unnecessary EAT and facilitate antibiotic stewardship.
Numerous prior studies have presented EHR-based machine learning models and clinical decision support systems to predict infection related conditions, such as bacteremia, sepsis, and ICU mortality.45–56 The goal of such models has been to ensure all septic and/or bacteremic patients are identified and treated early with appropriate antibiotic regimens.46–49,52–54,56 For instance, Nemati et al. achieved AUROCs ranging from 0.83–0.85 in predicting the early onset of sepsis using data collected during the 12, 8, 6, and 4 hours prior to diagnosis for patients across two Emory University hospitals and the MIMIC-III dataset. In contrast to these prior studies, the models we present differ by clinical timeframe (it is intended to be used after a patient is already suspected of having BI and has started EAT) and by the goal of the model (identify patients on EAT who are candidates for EAT discontinuation). Currently, no other prominent EHR-based prediction models exist with the goal of identifying patients on EAT with low risk of having BI who are candidates for EAT discontinuation. Existing methods for forecasting patient-level BI risk have focused around the use of protein and genetic biomarkers.6,41,42 The models we present rely on data commonly recorded in the ICU and do not require any specialized laboratory diagnostics or data from current BI risk prediction methods. Our study adds to the body of research surrounding EHR-based prediction models and provides a complementary approach to biomarker-based forecasting of patient-level BI risk. When used in combination with current BI risk metrics and clinical intuition, our model promises to help assist care providers in the de-escalation process of antibiotic stewardship.
For our clinical use case, false negative patients, i.e. those with a serious BI who were predicted as unlikely to have an infection, encompass the largest source of potential patient harm given the risk of untreated BIs in the ICU and therefore need to be minimized. Similarly, the largest source of potential patient benefit of our model from the current standard of care comes from reducing the number of antibiotic days given to patients who don’t have known BI. Our T=24-hour Random Forests model uses a high sensitivity decision threshold to in order to reduce false negative predictions and therefore improve the potential clinical utility in an ICU setting.
We recognize several limitations of this study. First, the retrospective data used was collected for clinical care purposes at a single academic medical center. The retrospective design of our study required us to infer information regarding BI suspicion, consecutive antibiotic days, and culture results based upon sensible criteria that may not completely reflect real world conditions. To address this, chart review and a variety of other quality checks were performed throughout the workflow to ensure appropriate coding of outcomes. Results from our 10-patient chart review of unknown BI status patients found two indeterminate cases and zero misclassifications by our proposed model. Details in the chart notes of one of these indeterminate cases suggested that this patient experienced a prolonged stay in the emergency department prior to transferring to the ICU and that the data from the emergency department was not available in the MIMIC-III dataset. This case suggests that the performance of our phenotype and model can be improved with more complete data on patients prior to ICU transfer. Future work will include retrospective data from additional ICU centers for external model validation and assessment of clinical utility, including data prior to ICU admission. Next, our estimates of antibiotic reduction provide an upper and lower bound on the potential clinical impacts of our model and makes numerous assumptions. To better understand the clinical utility of our model, further study is necessary to test the hypothesis that discontinuing antibiotic therapy on the patients predicted as low risk of BI would clinically benefit them. In future work, we will perform a propensity-matched analysis to estimate the effects of receiving short vs. prolonged antibiotics on outcome in patients with a predicted low risk of BI. Finally, the longitudinal patient data collected over T=24-,48-, or 72-hours was aggregated prior to modeling using the aggregation function(s) most associated with increased BI risk for each variable. With this design, the time for patients to exhibit symptoms most indicative of BI risk increases as the data collection window increases; however, time-window aggregation methods do capture temporal patterns present in the data to the fullest extent. To better leverage the longitudinal nature of our data, future work will focus on testing more complex algorithms to explore temporal trends and improve model performances.
5. Conclusion
The goal of this paper was to detail the design and initial application of a novel collection of algorithms which extract patient features from clinical data and identify patients at low risk of BI who can be safely removed from EAT at 24-hours after initiation. Our models achieved up to 0.8 AUC and demonstrate the feasibility of forecasting BI risk in a critical care setting using patient features found in the EHR. Future work will focus on validating models with external datasets, measuring clinical utility more accurately, and improving model performance by accounting for temporal information in patient data. Overall, these results call for more extensive research in this promising, yet relatively understudied, area.
Supplementary Material
Highlights.
Unnecessary antibiotic regimens can harm patients without bacterial infections.
Our random forest-based prediction model can help predict bacterial infection risk.
Data-driven approaches can enhance antibiotic stewardship efforts.
6. Acknowledgment
LNSP and YL are co-corresponding authors. This research is partly supported by R21 LM012618 (Luo), grant 5T32LM012203-04 from NLM (Eickelberg), and R21 LM012618 from NICHD (Sanchez-Pinto).
Glossary
- ICU
Intensive Care Unit
- BI
Bacterial Infection
- EAT
Empiric Antibiotic Therapy
- EHR
Electronic Health Record
- MIMIC-III
Medical Information Mart for Intensive Care III dataset
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
* The authors have no competing interests to declare
Contributor Information
Nelson Sanchez-Pinto, Department of Pediatrics (Critical Care) and Preventive Medicine (Health & Biomedical Informatics), 225 E. Chicago Avenue, Chicago, IL, 60611.
Yuan Luo, Department of Preventitive Medicine (Health & Biomedical Informatics), Feinberg School of Medicine, 750 N Lake Shore, Chicago, IL, 60611 E..
7. References
- 1.Claridge JA, Pang P, Leukhardt WH, Golob JF, Carter JW, Fadlalla AM. Critical analysis of empiric antibiotic utilization: establishing benchmarks. Surgical infections. 2010;11(2):125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francino MP. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Frontiers in Microbiology. 2015;6:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luyt C-E, Bréchot N, Trouillet J-L, Chastre J. Antibiotic stewardship in the intensive care unit. Critical Care. 2014;18(5):480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas Z, Bandali F, Sankaranarayanan J, Reardon T, Olsen KM. A Multicenter Evaluation of Prolonged Empiric Antibiotic Therapy in Adult ICUs in the United States. Critical care medicine. 2015;43(12):2527–2534. [DOI] [PubMed] [Google Scholar]
- 5.Weiss CH, Persell SD, Wunderink RG, Baker DW. Empiric antibiotic, mechanical ventilation, and central venous catheter duration as potential factors mediating the effect of a checklist prompting intervention on mortality: an exploratory analysis. BMC health services research. 2012;12:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zilahi G, McMahon MA, Povoa P, Martin-Loeches I. Duration of antibiotic therapy in the intensive care unit. Journal of Thoracic Disease. 2016;8(12):3774–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arulkumaran N, Routledge M, Schlebusch S, Lipman J, Conway Morris A. Antimicrobial-associated harm in critical care: a narrative review. Intensive care medicine. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surveillance of Antimicrobial Resistance in Europe. In: Control ECfDPa, ed 2017. [Google Scholar]
- 9.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infection and Drug Resistance. 2019;Volume 12:3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antibiotic resistance threats in the United States, 2013. In: Prevention CfDCa, Services UDoHaH, eds 2013. [Google Scholar]
- 12.More evidence on link between antibiotic use and antibiotic resistance. ScienceDaily: European Centre for Disease Prevention and Control (ECDC); 07/27/2017 2017. [Google Scholar]
- 13.Antimicrobial resistance: global report on surveillance. World Health Organization;2014. [Google Scholar]
- 14.Shallcross LJ, Davies DSC. Antibiotic overuse: a key driver of antimicrobial resistance. Br J Gen Pract. 2014;64(629):604–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael CA, Dominey-Howes D, Labbate M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Frontiers in Public Health. 2014;2:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Core Elements of Hospital Antibiotic Stewardship Programs | Antibiotic Use | CDC. 2019.
- 17.Camins BC, King MD, Wells JB, et al. Impact of an antimicrobial utilization program on antimicrobial use at a large teaching hospital: a randomized controlled trial. Infect Control Hosp Epidemiol. 2009;30(10):931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell BG, Schellevis F, Stobberingh E, Goossens H, Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infectious Diseases. 2014;14(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goff DA, File TM. The risk of prescribing antibiotics “just-in-case” there is infection. Seminars in Colon and Rectal Surgery. 2018;29(1):44–48. [Google Scholar]
- 20.Vincent J-L, Rello J, Marshall J, et al. International Study of the Prevalence and Outcomes of Infection in Intensive Care Units. JAMA. 2009;302(21):2323–2329. [DOI] [PubMed] [Google Scholar]
- 21.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical care medicine. 2001;29(7):1303–1310. [DOI] [PubMed] [Google Scholar]
- 22.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5(1):4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent JL, Abraham E, Annane D, Bernard G, Rivers E, Van den Berghe G. Reducing mortality in sepsis: new directions. Crit Care. 2002;6 Suppl 3:S1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincent JL, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. Jama. 2009;302(21):2323–2329. [DOI] [PubMed] [Google Scholar]
- 25.Andre K, Mark M, Michael K, et al. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. American journal of respiratory and critical care medicine. 2005;171(4):388–416. [DOI] [PubMed] [Google Scholar]
- 26.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Critical care medicine. 2013;41(2):580–637. [DOI] [PubMed] [Google Scholar]
- 27.Solomkin JS, Mazuski JE, Bradley JS, et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2010;50(2):133–164. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman JJ. Society of Critical Care Medicine Presidential Address−47th Annual Congress, February 2018, San Antonio, Texas. Critical care medicine. 2018;46(6):839–842. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Critical care medicine. 2006;34(6):1589–1596. [DOI] [PubMed] [Google Scholar]
- 30.Misquitta D. Early Prediction of Antibiotics in Intensive Care Unit Patients [Master’s Thesis]: Biomedical Informatics, Harvard Medical School; 2013. [Google Scholar]
- 31.Luo Y, Szolovits P, Dighe AS, Baron JM. 3D-MICE: integration of cross-sectional and longitudinal imputation for multi-analyte longitudinal clinical data. (1527–974X (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Szolovits P, Dighe AS, Baron JM. Using Machine Learning to Predict Laboratory Test Results. (1943–7722 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 33.Le Cessie S, Van Houwelingen JC. Ridge Estimators in Logistic Regression. Journal of the Royal Statistical Society Series C (Applied Statistics). 1992;41(1):191–201. [Google Scholar]
- 34.Breiman L Random Forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 35.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20(3):273–297. [Google Scholar]
- 36.Friedman JH. Stochastic gradient boosting. Computational Statistics & Data Analysis. 2002;38(4):367–378. [Google Scholar]
- 37.Luna C, Blanzaco D, Niederman M, et al. Resolution of ventilator-associated pneumonia: Prospective evaluation of the clinical pulmonary infection score as an early clinical predictor of outcome*. Critical care medicine. 2019;31(3):676–682. [DOI] [PubMed] [Google Scholar]
- 38.BLOT F, RAYNARD B, CHACHATY E, TANCRÈDE C, ANTOUN S, NITENBERG G. Value of Gram Stain Examination of Lower Respiratory Tract Secretions for Early Diagnosis of Nosocomial Pneumonia. http://dxdoiorg/101164/ajrccm16259908088 2000. [DOI] [PubMed]
- 39.Campion M, Scully G. Antibiotic Use in the Intensive Care Unit: Optimization and De-Escalation. J Intensive Care Med. 2018;33(12):647–655. [DOI] [PubMed] [Google Scholar]
- 40.Samuel LP, Balada-Llasat J-M, Harrington A, Cavagnolo R, Bourbeau P. Multicenter Assessment of Gram Stain Error Rates. 2016. [DOI] [PMC free article] [PubMed]
- 41.de Jong E, van Oers JA, Beishuizen A, et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. The Lancet Infectious Diseases. 2016;16(7):819–827. [DOI] [PubMed] [Google Scholar]
- 42.Schuetz P, Wirz Y, Sager R, et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. The Cochrane database of systematic reviews. 2017;10:Cd007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cals JW, Ebell MH. C-reactive protein: guiding antibiotic prescribing decisions at the point of care. Br J Gen Pract. 2018;68(668):112–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paonessa JR, Shah RD, Pickens CI, et al. Rapid Detection of Methicillin-Resistant Staphylococcus aureus in BAL: A Pilot Randomized Controlled Trial. Chest. 2019;155(5):999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward L, Paul M, Andreassen S. Automatic learning of mortality in a CPN model of the systemic inflammatory response syndrome. Math Biosci. 2017;284:12–20. [DOI] [PubMed] [Google Scholar]
- 46.Ward L, Møller JK, Eliakim-Raz N, Andreassen S. Prediction of Bacteraemia and of 30-day Mortality Among Patients with Suspected Infection using a CPN Model of Systemic Inflammation. IFAC-PapersOnLine. 2018;51(27):116–121. [Google Scholar]
- 47.Parente JD, Möller K, Shaw GM, Chase JG. Hidden Markov Models for Sepsis Classification. IFAC-PapersOnLine. 2018;51(27):110–115. [Google Scholar]
- 48.Paul M, Andreassen S, Nielsen AD, et al. Prediction of bacteremia using TREAT, a computerized decision-support system. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2006;42(9):1274–1282. [DOI] [PubMed] [Google Scholar]
- 49.Sheetrit E, Nissim N, Klimov D, Shahar Y. Temporal Probabilistic Profiles for Sepsis Prediction in the ICU. Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining; 2019; Anchorage, AK, USA. [Google Scholar]
- 50.Vieira SM, Carvalho JP, Fialho AS, Reti SR, Finkelstein SN, Sousa JMC. A Decision Support System for ICU Readmissions Prevention. Proceedings of the 2013 Joint Ifsa World Congress and Nafips Annual Meeting (Ifsa/Nafips) 2013:251–256. [Google Scholar]
- 51.Luo Y, Xin Y, Joshi R, Celi L, Szolovits P. Predicting ICU mortality risk by grouping temporal trends from a multivariate panel of physiologic measurements. Paper presented at: Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence; 02/12/2016, 2016. [Google Scholar]
- 52.Gultepe E, Green JP, Nguyen H, Adams J, Albertson T, Tagkopoulos I. From vital signs to clinical outcomes for patients with sepsis: a machine learning basis for a clinical decision support system. In: J Am Med Inform Assoc. Vol 21 2014:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brause R, Hamker F, Paetz J. Septic Shock Diagnosis by Neural Networks and Rule Based Systems In: Computational intelligence techniques in medical diagnosis and prognosis. SpringerLink; 2002. [Google Scholar]
- 54.Peelen L, de Keizer NF, Jonge E, Bosman RJ, Abu-Hanna A, Peek N. Using hierarchical dynamic Bayesian networks to investigate dynamics of organ failure in patients in the Intensive Care Unit. J Biomed Inform. 2010;43(2):273–286. [DOI] [PubMed] [Google Scholar]
- 55.Curto S, Carvalho JP, Salgado C, Vieira SM, Sousa JMC. Predicting ICU readmissions based on bedside medical text notes. Paper presented at: 2016 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE); 24–29 July 2016, 2016. [Google Scholar]
- 56.Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Critical care medicine. 2018;46(4):547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


