Figure 1.
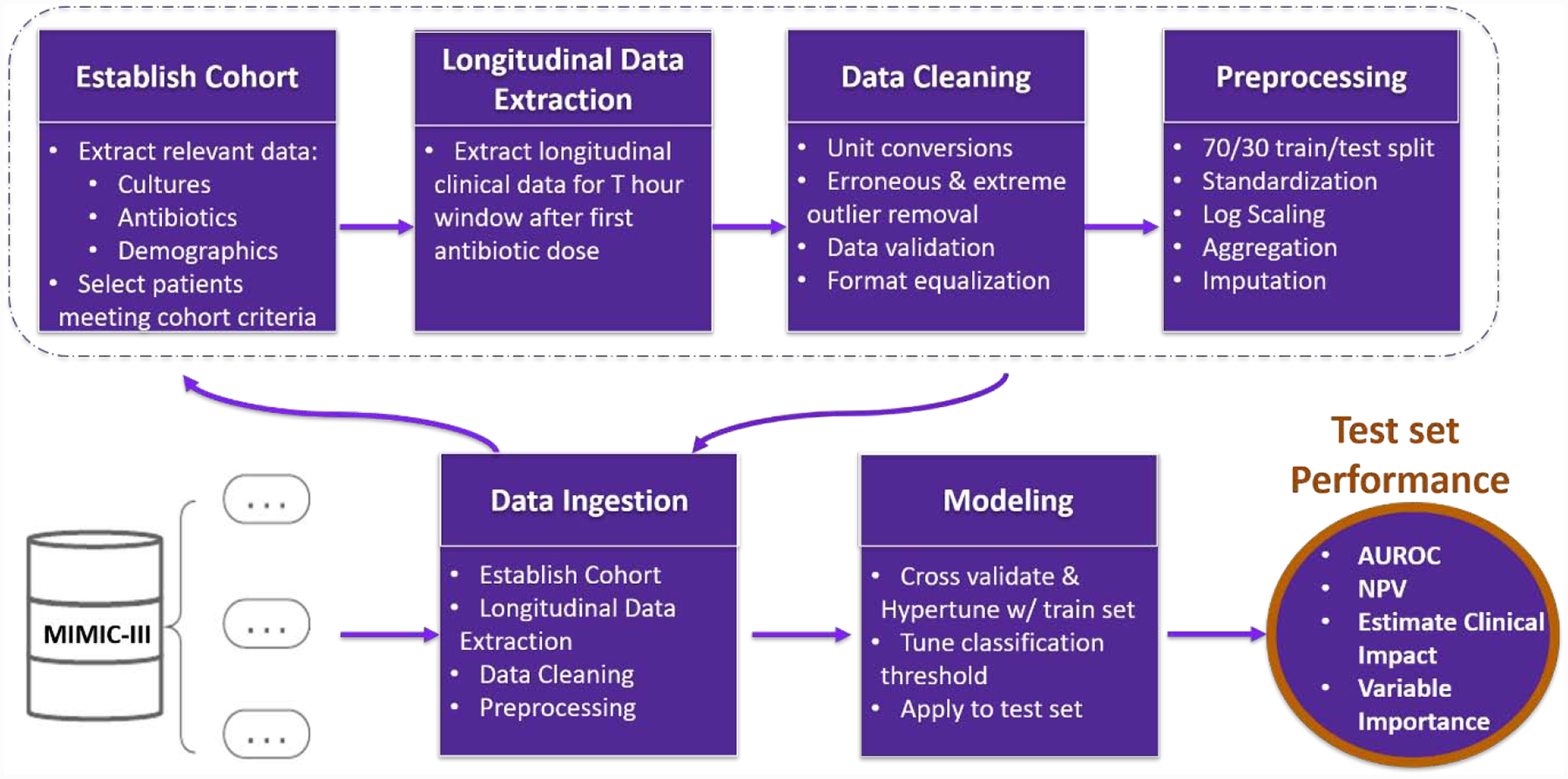
Data Ingestion and Analysis Framework Overview. Raw data is ingested from the MIMIC-III database. First a cohort of adult patients suspected of having SBI is established, and both longitudinal and categorical data is extracted over the T= 24-,48-, or 72-hour window following their first antibiotic dose that corresponds with an microbiologic culture. Next, data is cleaned, formatted, and preprocessed prior to modeling. The cohort is then filtered to patients with positive microbiologic culture and prolonged antibiotics, and microbiologic culture negative with short antibiotics. A 70/30 train/test set split is then applied. Scaling and standardization are performed on each set independently. Missing values were imputed using median values from the training set. Machine learning models are hypertuned on the training set and applied to the test set. Finally, classification thresholds are tuned, and model performance metrics are output.
