Abstract
Cervicovaginal secretions, or their components collected, are referred to as cervicovaginal lavage (CVL). CVL constituents have utility as biomarkers and play protective roles in wound healing and against HIV‐1 infection. However, several components of cervicovaginal fluids are less well understood, such as extracellular RNAs and their carriers, for example, extracellular vesicles (EVs). EVs comprise a wide array of double‐leaflet membrane extracellular particles and range in diameter from 30 nm to over one micron. The aim of this study was to determine whether differentially regulated CVL microRNAs (miRNAs) might influence retrovirus replication. To this end, we characterized EVs and miRNAs of primate CVL during the menstrual cycle and simian immunodeficiency virus (SIV) infection of macaques. EVs were enriched by stepped ultracentrifugation, and miRNA profiles were assessed with a medium‐throughput stem‐loop/hydrolysis probe qPCR platform. Whereas hormone cycling was abnormal in infected subjects, EV concentration correlated with progesterone concentration in uninfected subjects. miRNAs were present predominantly in the EV‐depleted CVL supernatant. Only a small number of CVL miRNAs changed during the menstrual cycle or SIV infection, for example, miR‐186‐5p, which was depleted in retroviral infection. This miRNA inhibited HIV replication in infected macrophages in vitro. In silico target prediction and pathway enrichment analyses shed light on the probable functions of miR‐186‐5p in hindering HIV infections via immunoregulation, T‐cell regulation, disruption of viral pathways, etc. These results provide further evidence for the potential of EVs and small RNAs as biomarkers or effectors of disease processes in the reproductive tract.
Keywords: cervicovaginal lavage, exosome, extracellular vesicle, HIV‐1, microRNA
By comparing the microRNA and extracellular vesicle profiles of cervicovaginal lavage of both uninfected and infected macaques, we observed an enrichment of microRNAs in extracellular vesicle‐depleted supernatant and discovered a significant reduction in miR‐186‐5p after simian immunodeficiency virus infection. Furthermore, once transfected into HIV‐infected human macrophages, synthetic miR‐186‐5p mimics inhibited HIV replication, demonstrating its antiretroviral potential.
![]()
Abbreviations
- ACD
acid citrate dextrose
- CVL
cervicovaginal lavage
- EV
extracellular vesicle
- exRNAs
extracellular RNAs
- exRNPs
extracellular ribonucleoprotein complexes
- IFN‐γ
interferon‐γ
- M‐CSF
macrophage colony‐stimulating factor
- MDM10
macrophage differentiation medium with 10% FBS
- MDM20
macrophage differentiation medium with 20% FBS
- miRNAs
microRNAs
- NTA
nanoparticle tracking analysis
- PBMCs
peripheral blood mononuclear cells
- RBC
red blood cell
- SIV
simian immunodeficiency virus
- TLDA
TaqMan low‐density array
- UC
ultracentrifuge
The cervicovaginal canal is a potential source of biological markers for forensics investigations [1, 2, 3, 4], reproductive tract cancers [5, 6, 7], and infections [8, 9, 10]. Cervicovaginal secretions may be collected by swab, tampon, or other methods, or secretion components may be liberated by a buffered wash solution and collected as cervicovaginal lavage (CVL). In addition to utility as biomarkers, constituents of cervicovaginal secretions, including proteins, certain microbes, and metabolites, exert function, for example, by playing protective roles in wound healing [11] and against HIV‐1 infection [12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22]. A large and important body of work has thus examined biomarker potential and functional roles of numerous entities in the cervicovaginal compartment.
Compared with secreted proteins, metabolites, and the microbiome, however, several components of cervicovaginal fluids are less well understood, including extracellular RNAs (exRNAs) and their carriers, such as extracellular vesicles (EVs) and extracellular ribonucleoprotein complexes (exRNPs). EVs are potential regulators of cell behavior in paracrine and endocrine fashion due to their reported abilities to transfer proteins, nucleic acids, sugars, and lipids between cells [23]. EVs comprise a wide array of double‐leaflet membrane extracellular particles, including those of endosomal and cell‐surface origin [24, 25], and range in diameter from 30 nm to well over one micron (large oncosomes) [26]. EV macromolecular composition tends to reflect, but is not necessarily identical to, that of the cell of origin [27]. EVs have been isolated from most cells, as well as biological fluids [23, 28], including cervicovaginal secretions of humans [29] and rhesus macaques [30].
MicroRNAs (miRNAs) are one of the most studied classes of exRNA. These noncoding RNAs average 22 nucleotides in length and, in some cases, fine‐tune the expression of target transcripts [31, 32]. Released from cells by several routes, miRNAs are among the most frequently examined biomarker candidates in biofluids and, along with some other RNAs, are reported to be transmitted via EVs [33, 34, 35, 36]. miRNAs are found not only in EVs, but also in free Argonaute‐containing protein complexes; the latter may outnumber the former, at least in blood [37, 38]. Many miRNAs are also highly conserved [32], and abundant species typically have 100% identity in humans and nonhuman primates [39]. [For this reason, we will refer to hsa‐ (Homo sapiens) and mml‐ (Macaca mulatta) miRNAs without the species designation unless otherwise warranted by sequence disparity.] While miRNAs have been profiled in cervicovaginal secretions and menstrual blood, mostly in the forensics setting [4, 40, 41], their associations with EV and exRNP fractions require further study. A recent publication reported that EVs from healthy vaginal secretions inhibited HIV‐1 infection [29]. Another report found that CVL EVs (styled “exosomes”) were present at higher concentrations in cervical cancer and that two miRNAs were also upregulated [5]. Our laboratory described a reduction of CVL EVs in a severe endometriosis case compared with reproductively healthy primates [30]. However, our study, along with others, was limited by the absence of molecular profiling of EV cargo [30].
Many immunocompetent cell types populate the female reproductive tracts, among which are vaginal‐resident macrophages. These cells reside in the lamina propria and participate in the host's innate immune responses via specialized phagocytic elimination and limited antigen‐presenting capability [42, 43]. Once activated, for example, by sensing interferon‐γ (IFN‐γ) from Th1 effectors cells [43], vaginal macrophages increase their degradative ability. The recruitment of vaginal macrophages is sex hormone‐dependent, and their phagocytotic capability is not impaired by the low pH of the microenvironment [43, 44]. Numerous papers have demonstrated that CD68+ macrophages express receptors CD4, CCR5, and CXCR4, indicating that they are susceptible to infection by both R5 and X4‐tropic HIV‐1 virus during genital infection and transmission [42, 43, 45, 46, 47].
Here, we performed targeted miRNA profiling of EV‐enriched and EV‐depleted fractions of CVL and vaginal secretions collected from healthy and retrovirus‐infected rhesus macaques. We queried how CVL EVs and miRNAs are affected by the menstrual cycle, an important potential confounder of biomarker studies [48]. Similarly, we assessed possible associations with simian immunodeficiency virus (SIV) infection. We report an association of miR‐186 levels with SIV infection and find that this miRNA also appears to have antiretroviral effects in HIV‐infected macrophages. These studies provide baseline information for easily accessed CVL markers including EVs and miRNAs that may become useful tools in the clinic.
Materials and methods
Sample collection
Cervicovaginal lavage and whole blood samples were collected weekly for 5 weeks from two uninfected (control) and four SIVmac251‐infected (infected) rhesus macaques (M. mulatta) as previously described [30]. All macaques were negative for simian T‐cell leukemia virus and simian type D retrovirus and were inoculated intravenously. Animals were sedated with ketamine at a dose of 7–10 mg·kg−1 prior to all procedures. CVL was performed by washing the cervicovaginal cavity with 3 mL of PBS (Thermo Fisher Scientific, Waltham, MA, USA. Cat #: 14190‐144) directed into the cervicovaginal canal and re‐aspirated using the same syringe. Materials and procedures for sample collection are depicted in Fig. S1. Volumes of CVL yield across collection dates were documented in Table S1. Whole blood (3 mL) was collected by venipuncture into syringes containing acid citrate dextrose (ACD) solution (Sigma‐Aldrich, St. Louis, MO, USA. Cat #: C3821).
Study approvals
All animal studies were approved by the Johns Hopkins University Institutional Animal Care and Use Committee and conducted in accordance with the Weatherall Report, the Guide for the Care and Use of Laboratory Animals, and the USDA Animal Welfare Act.
Sample processing
Sample processing began within a maximum of 60 min of collection and utilized serial centrifugation steps to enrich EVs as described previously [30], based on a standard EV isolation protocol [49]. Specifically, fluids were centrifuged: (a) 1000 g for 15 min at 4 °C in a tabletop centrifuge; (b) 10 000 g for 20 min at 4 °C; and (c) 110 000 g for 2 h at 4 °C with a Sorvall Discovery SE ultracentrifuge (Thermo Fisher Scientific) with an AH‐650 rotor (k factor: 53.0; Fig. S1B). Following each centrifugation step, most supernatant was removed, taking care not to disturb the pellet. After each step, supernatant was set aside for nanoparticle tracking analysis (NTA; 200 µL) and RNA isolation (200 µL) following the second and third steps. The pellet was resuspended in 400 µL of PBS after each centrifugation step. After the final step, the remaining ultracentrifuged (UC) supernatant was concentrated to approximately 220 µL using Amicon Ultra‐2 10‐kDa molecular weight cutoff filters (Merck KGaA, Darmstadt, Germany. Cat #: UFC201024). Two hundred microliter of the concentrate was used for RNA isolation, and the remainder was retained for NTA. All samples reserved for RNA isolation were mixed with 62.6 µL of RNA isolation buffer (Exiqon, Vedbaek, Denmark. Cat #: 300112. Lot #: 593‐84‐9n) containing three micrograms of glycogen and 5 pg of synthetic cel‐miR‐39 as previously described [50]. Processed samples were analyzed immediately or frozen at −80 °C until further use.
For plasma, whole blood was centrifuged at 800 g for 10 min at 25 °C. Supernatant was centrifuged twice at 2500 g for 10 min at 25 °C. The resulting platelet‐poor plasma was aliquoted and frozen at −80 °C.
Hormone analysis
Levels of progesterone (P4) and estradiol‐17b (E2) were measured in plasma samples shipped overnight on dry ice to the Endocrine Technology and Support Core Lab at the Oregon National Primate Research Center, Oregon Health and Science University.
Nanoparticle tracking analysis
Extracellular particle concentration was determined using a NanoSight NS500 NTA system (Malvern, Worcestershire, UK). CVL samples were diluted as needed and specified in Table S2 to ensure optimal NTA analysis. At least five 20‐s videos were recorded for each sample at a camera setting of 12. Data were analyzed at a detection threshold of two using nanosight software version 3.0.
Western blot
Western blot was used to detect the presence of EV protein markers and the absence of nucleoporin (nuclear marker) in CVL and enriched CVL EVs. Twenty microliter of samples from each fraction was lysed with 5 µL 1 : 1 mixture of RIPA buffer (Cell Signaling Technology, Danvers, MA, USA. Cat #: 9806S) and protease inhibitor (Santa Cruz Biotechnology, Dallas, TX, USA. Cat #: sc29131). Eight microliter of Laemmli 4× sample buffer (Bio‐Rad, Hercules, CA, USA. Cat #:161‐0747 Lot #: 64077737) was added per sample, and 30 µL of each was loaded into a Criterion TGX 4–15% gel (Bio‐Rad. Cat #: 5678084 Lot #: 64301319) after 5 min of 95 °C incubation. The gel was electrophoresed by application of 100 V for 100 min. The proteins were then transferred to a PVDF membrane (Bio‐Rad. Cat #: 1620177, Lot #:31689A12.), which was blocked with 5% milk (Bio‐Rad. Cat #: 1706404. Lot #: 64047053) in PBS + 0.1%Tween®20 (Sigma‐Aldrich, Cat #: 274348 Lot #: MKBF5463V) for 1 h. The membrane was subsequently incubated with mouse anti‐human CD63 (BD Biosciences, San Jose, CA, USA, Cat #: 556019 Lot #: 6355939) and mouse monoclonal IgG_2b CD81 (Santa Cruz Biotechnology, Cat #: 166029 Lot #: L1015) primary antibodies, at a concentration of 0.5 µg·mL−1 overnight. After washing the membrane, it was incubated with a goat anti‐mouse IgG‐HRP secondary antibody (Santa Cruz Biotechnology, Cat #: sc‐2005 Lot #: B1616) at a 1 : 5000 dilution for 1 h. The membrane was then incubated with a 1 : 1 mixture of SuperSignal West Pico Stable Peroxide solution and Luminol Enhancer solution (Thermo Scientific, Rockford, IL, USA, Cat #: 34080 Lot #: SD246944) for 5 min. The membrane was visualized on Azure 600 imaging system (Azure Biosystems, Dublin, CA, USA). The second blot was done in a reducing environment using 10 mm DTT (Promega, Madison, WI, USA, Cat #: P1171 Lot #: 0000198991). Same procedures were followed with rabbit anti‐human TSG101 (Cat #: ab125011 Lot #:GR180132‐14), rabbit polyclonal antinucleoporin (Abcam, Cambridge, MA, USA, Cat #: ab96134 Lot #: GR22167‐18) primary antibodies. Subsequent incubation with goat anti‐rabbit IgG‐HRP secondary antibody (Abcam, Cat #: sc‐2204 Lot #: B2216). All antibodies were used at the same concentration as the first blot. Membrane was visualized on the Azure imaging system.
Single‐particle interferometric reflectance imaging
Both CVL‐derived and dendritic cell LK23‐derived EVs were diluted 1 : 1000 and incubated on ExoView (NanoView Biosciences, Brighton, MA, USA) chips that were printed with anti‐CD63 (BD Biosciences, Cat#: 556019) and anti‐CD81 (BD Biosciences, Cat #: 555675) antibodies. After incubation for 16 h, chips were washed as per the manufacturer's protocol and imaged in the ExoView scanner by interferometric reflectance imaging.
Electron microscopy
Gold grids were floated on 2% paraformaldehyde‐fixed CVL‐derived samples for 2 min and then negatively stained with uranyl acetate for 22 s. Grids were observed with a Hitachi 7600 transmission electron microscope in the Johns Hopkins Institute for Basic Biomedical Sciences Microscope Facility.
Total RNA isolation and quality control
RNA isolation workflow is shown in Fig. S1C. RNA lysis buffer was added into each sample as described above prior to freezing (−80 °C). Total RNA was isolated from thawed samples using the miRCURY RNA Isolation Kit‐Biofluids (Exiqon, Cat #: 300112. Lot #: 593‐84‐9n) as per the manufacturer's protocol with minor modifications as previously described [50]. Total RNA was eluted with 50 µL RNase‐free water and stored at −80 °C. As quality control, expression levels of several small RNAs (sRNAs) including snRNA U6, miR‐16‐5p, miR‐223‐3p, and the spiked‐in synthetic cel‐miR‐39 were assessed by TaqMan miRNA assays (Applied Biosystems/Life Technologies, Carlsbad, CA, USA) [51].
miRNA profiling by TaqMan low‐density array
A custom 48‐feature TaqMan low‐density array (TLDA) was ordered from Thermo Fisher, with features chosen based on results of a human CVL pilot study (GVH and KWW, unpublished data). Stem‐loop primer reverse transcription and preamplification steps were conducted using the manufacturer's reagents as previously described [52] but with 14 cycles of preamplification. Real‐time quantitative PCR was performed with a QuantStudio 12K instrument (Johns Hopkins University DNA Analysis Facility). Data were collected using sds software and C q values extracted with expression suite v1.0.4 (Thermo Fisher Scientific). Raw C q values were adjusted by a factor determined from the geometric mean of 10 relatively invariant miRNAs. Normalizing to the geometric mean of multiple carefully selected housekeeping miRNAs is a biologically relevant and recommended normalization method [53]. The selection process for these invariant miRNAs was to (a) rank miRNAs by coefficient of variation; (b) remove miRNAs with high average C q (> 30), non‐miRNAs, and those with low amplification score; (c) select the lowest‐CV member of miRNA families (e.g., the 17/92 clusters); and (d) pick the top 10 remaining candidates by CV: let‐7b‐5p, ‐miR‐21‐5p, ‐27a‐3p, ‐28‐3p, ‐29a‐3p, ‐30b‐5p, ‐92a‐3p, ‐197‐3p, ‐200c‐3p, and ‐320a‐3p.
Individual RT‐qPCR assays
Individual TaqMan miRNA qPCR assays were performed as previously described [52] on all UC pellet samples from all animals across all weeks for miRs‐19a‐3p (Thermo Fisher Assay ID #000395), ‐186‐5p (Thermo Fisher Assay ID #002285), ‐451a‐5p (Thermo Fisher Assay ID #001105), ‐200c‐3p (Thermo Fisher Assay ID #002300), ‐222‐3p (Thermo Fisher Assay ID #002276), ‐193b‐3p (Thermo Fisher Assay ID #002367), ‐181a‐5p (Thermo Fisher Assay ID #000480), ‐223a‐3p (Thermo Fisher Assay ID #002295), ‐16‐5p (Thermo Fisher Assay ID #000391), ‐106a‐5p (Thermo Fisher Assay ID #002169), and ‐125b‐5p (Thermo Fisher Assay ID #00449). We also measured miR‐375‐3p (Thermo Fisher Assay ID #00564), which was not included on the array. Data were adjusted to Cqs of miR‐16‐5p.
Blood cell isolation and monocyte‐derived macrophage culture
Total peripheral blood mononuclear cells (PBMCs) were obtained from freshly drawn blood from human donors under a Johns Hopkins University School of Medicine IRB‐approved protocol (JHU IRB #CR00011400). Blood was mixed with 10% ACD (Sigma‐Aldrich, Cat #: C3821 Lot #: SLBQ6570V) with gentle mixing by inversion. Within 15 min of draw, blood was diluted with equal volume of PBS + 2% FBS and gently layered onto room temperature Ficoll (Biosciences AB, Uppsala, Sweden, Cat #:17‐1440‐03 Lot #: 10253776) in Sepmate‐50 tubes (STEMCELL Technologies, Vancouver, BC, Canada, Cat #: 15450 Lot #: 06102016) and centrifuged for 10 min at 1200 g. Plasma and PBMC fractions were removed, washed in PBS + 2% FBS, and pelleted at 300 g for 8 min. Pellets from five tubes were combined by resuspension in 10 mL red blood cell (RBC) lysis buffer (4.15 g NH4Cl, 0.5 g KHCO3, 0.15 g EDTA in 450 mL H2O; pH adjusted to 7.2–7.3; volume adjusted to 500 mL and filter‐sterilized); total volume was brought to 40 mL with RBC lysis buffer. After incubation at 37 °C for 5 min, the suspension was centrifuged at 400 g for 6 min at room temperature. The cell pellet was resuspended in macrophage differentiation medium with macrophage colony‐stimulating factor (M‐CSF) and 20% FBS (MDM20) to a final concentration of 2 × 106 cells·mL−1. PBMCs were plated at 4 × 106 cells per well in 12‐well plates and cultured in MDM20 for 7 days. One half of the total volume of medium was replaced on day 3. On day 7, cells were washed three times with PBS to remove nonadherent cells. The medium was replaced with macrophage differentiation medium with M‐CSF and 10% serum (MDM10) and cultured overnight prior to transfection.
miRNA mimic transfection
Differentiated macrophages were transfected with 50 nm miRNA‐186‐5p (Qiagen, Foster City, CA, USA. Cat #: MSY0000456 Lot #: 286688176) using Lipofectamine 2000 (Invitrogen/Life Technologies, Carlsbad, CA, USA, Cat #: 11668‐019 Lot #:1467572) diluted in OptiMEM Reduced Serum Medium (Gibco, Grand Island, NY, USA, Cat #: 31985‐070 Lot #: 1762285). Controls included mock transfections and transfection of 50 nm double‐stranded siRNA oligo labeled with Alexa Fluor 555 (Invitrogen, Fredrick, MD, USA, Cat #: 14750‐100 Lot #: 1863892). Plates were incubated for 6 h at 37 °C. After incubation, successful transfection was confirmed by examining uptake of labeled siRNA with an Eclipse TE200‐inverted microscope (Nikon Instruments, Melville, NY, USA). Transfection medium was removed. The plates were washed with PBS and refed with 2 mL fresh MDM10 medium.
HIV infection
HIV‐1 BaL stocks were generated from infected PM1 T lymphocytic cells and stored at −80 °C. 24 h after mimic or mock transfections; macrophages were infected with HIV BaL and incubated overnight (stock, 80 μg p24·mL−1, diluted to 200 ng p24·mL−1). At days 3, 6, and nine postinfection, 500 μL supernatant was collected for p24 release assays and cells were lysed with 600 μL mirVana lysis buffer for subsequent RNA isolation and analysis.
HIV p24 antigen ELISA
Supernatant samples were lysed with Triton X (Perkin Elmer, Waltham, MA, USA, Cat #: NEK050B001KT Lot #: 990‐17041) at a final concentration of 1%. The DuPont HIV‐1 p24 Core Profile ELISA kit (Perkin Elmer, Cat #: NEK050B001KT Lot #: 990‐17041) was used as per the manufacturer's instructions to measure p24 concentration based on the provided standard.
Total RNA isolation
Total RNA was isolated using the mirVana miRNA Isolation Kit as per the manufacturer's protocol (Ambion, Vilnius, Lithuania. Cat #: AM1560 Lot #: 1211082). Note that this procedure yields total RNA, not just sRNAs. After elution with 100 μL RNase‐free water, nucleic acid concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). RNA isolates were stored at −80 °C.
HIV Gag RNA RT‐qPCR
Real‐time one‐step reverse transcription–quantitative PCR was performed with the QuantiTect Virus Kit (Qiagen, Cat #:211011 Lot #: 154030803). Each 25 μL reaction mixture contained 15 μL of master mix containing HIV‐1 RNA standard, 100 μm of FAM dye, and IBFQ quencher labeled Gag probe (5′ ATT ATC AGA AGG AGC CAC CCC ACA AGA 3′), 600 nm each of Gag1 forward primer (5′ TCA GCC CAG AAG TAA TAC CCA TGT 3′) and Gag2 reverse primer (5′ CAC TGT GTT TAG CAT GGT GTT T 3′), nuclease‐free water, and QuantiTect Virus RT mix, and 10 μL serial‐diluted standard or template RNA. No‐template control and no reverse transcriptase controls were included. Linear standard curve was generated by plotting the log copy number versus the quantification cycle (C q) value. Log‐transformed Gag copy number was calculated based on the standard curve.
Data analysis
Data processing and analysis were conducted using tools from Microsoft Excel (geometric mean normalization), Apple Numbers, GraphPad Prism, the MultiExperiment Viewer, and r/BioConductor packages including pheatmap (http://CRAN.R‐project.org/package=pheatmap; quantile normalization, Euclidean distance, self‐organizing maps, self‐organizing tree algorithms, k‐means clustering).
Results
Abnormal menstrual cycle of SIV‐infected macaques and ovulation‐associated changes in CVL EV‐enriched particles
Plasma and CVL were collected from two control and four SIV‐infected macaques over the course of 5 weeks (Fig. S1). Amenorrhea (absence of menstruation) was observed for infected subjects (K. Mulka, et al, unpublished data). By NTA, CVL EV concentration in control animals increased during ovulation (Fig. 1A). Transmission electron microscopy was performed for representative fractions of CVL, revealing bacteria and large particles in the 10 000 g pellet (Fig. 1B). The 100 000 g pellet included apparent EVs up to 200 nm in diameter (Fig. 1C). EV markers (shown: CD63, CD81, and TSG101) were confirmed by Western blot (Fig. 1D). The nuclear marker nucleoporin was detected only in tissue samples (Fig. 1D). The relative EV tetraspanin profiles of both CVL and control EV samples were corroborated with single‐particle interferometric reflectance imaging: CVL EVs had a higher CD63 expression, and dendritic cell EVs had higher CD81 expression.
Fig. 1.

EV composition during the menstrual cycle. (A) Nanoparticle concentrations of CVL UC pellets monitored weekly over 5 weeks for two SIV‐negative (“control”) and four SIV‐infected rhesus macaques (Mean ± SD). Red arrows indicate time of ovulation for two control animals, which were absent for SIV‐infected animals. (B) Transmission electron micrographs of CVL pellets from the 10 000 g pellet (left) and 110 000 g pellet (right) confirm presence of bacteria and EVs/EV‐like particles, with several respective diameters indicated. Scale bar = 500 nm. (C) Western blot analysis suggests enrichment of EV markers CD63, CD81, and TSG101 in 110k pellet fraction of CVL from uninfected animals (n = 2). Vaginal tissue homogenate and DC (LK23) 110k pellet controls were also positive for CD63 and CD81. Nuclear marker nucleoporin was detected in tissue homogenate but not in putative EV samples. Dashed lines were added to indicate the grouping of the ladder and the samples on the TSG101 and nucleoporin blots. (D) SP‐IRIS confirmation of EV markers on CVL and DC EVs. Shown are averages of tetraspanin‐positive particles bound to anti‐CD63 and anti‐CD81 antibodies and detected by label‐free imaging (mean ± SD).
TLDA reveals an extracellular miRNA profile of the cervicovaginal compartment
Based upon preliminary findings from a study of human CVL (Hancock and Witwer, unpublished data), we designed a custom TLDA to measure 47 miRNAs expected to be present in CVL, along with the snRNA U6. CVL from all subjects and at all time points was fractionated by stepped centrifugation to yield a 10 000 g pellet (10 K pellet), a 100 000 g pellet (UC pellet), and 100 000 g supernatant (UC supernatant). Total RNA from all fractions was profiled by TLDA. Raw (Fig. S2A), quantile normalized (Fig. S2B), and geometric mean‐adjusted C q values (Fig. S2C) were subjected to unsupervised hierarchical clustering. This clustering did not reveal broad miRNA profile differences associated with sample collection time, menstruation, or SIV infection.
Distribution of miRNAs across CVL fractions
Across the three examined CVL fractions (p10, p100, S100), the 10 most abundant miRNAs (lowest C q values) were miRs‐223‐3p, ‐203a‐3p, ‐24‐3p, ‐150‐5p, ‐21‐5p, ‐146a‐5p, ‐92a‐3p, ‐222‐3p, ‐17‐5p, and ‐106a‐5p. The average normalized C q value for each miRNA was greater (i.e., lower abundance) in the p100 than the s100 fraction (Fig. 2A and inset) and indeed in p10 and p100 combined (Fig. 2B), suggesting that most miRNA in CVL, as reported for various other body fluids, is found outside the EV‐enriched fractions. Considering all fractions, the differences between the EV‐enriched and EV‐depleted fractions were significant even after Bonferroni correction for all features except U6. On average, the s100 fraction contained 86.5% of the total miRNA from these three fractions. In the p10 fraction, the average miRNA was detected at 10.5% its level in the s100 fraction (SD = 5.7%). miR‐34a‐5p had the lowest (5.9%) and miR‐28‐3p the highest (33.7%) abundance compared with s100. In the p100 fraction, miRNAs were on average 5.6% (SD = 2.4%) as abundant as in s100. The least represented in p100 was miR‐27a‐3p (2.3%), and the best represented was again miR‐28‐3p (13.4%). Together, the content of the EV‐enriched fractions (p10 and p100) as a percentage of the total is shown in Fig. 2B for individual miRNAs. miRNA rank was significantly correlated across fractions, despite minor differences in order (Fig. 2C).
Fig. 2.
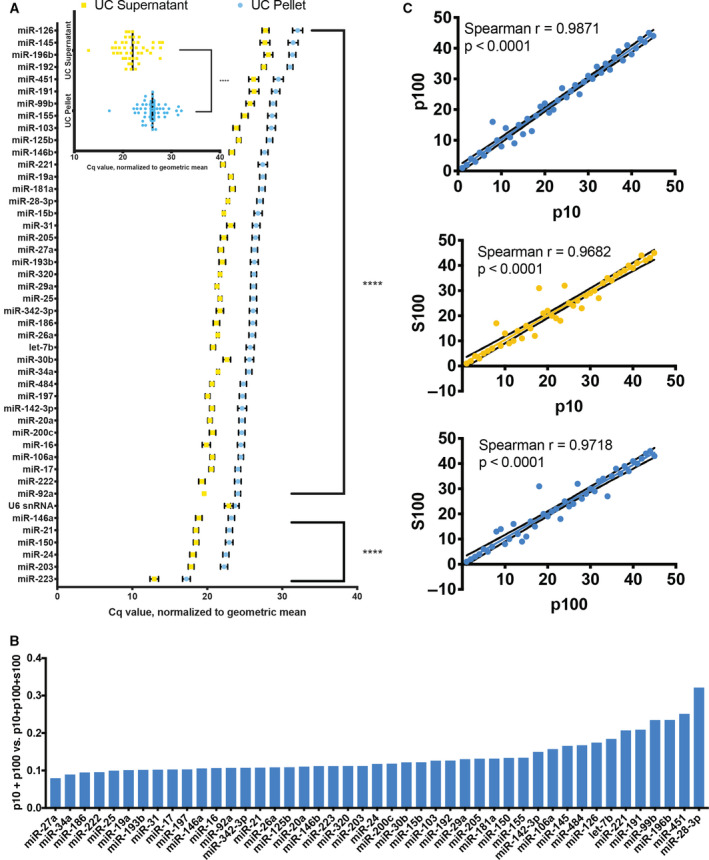
Relative abundance of miRNAs in different CVL fractions from all subjects (n = 6). (A) Abundant miRNAs in descending order based on C q values normalized to the geometric mean for each sample. Inset: average of all miRNAs in UC pellet and UC supernatant. Error bars: SEM. (B) miRNA expression in EV‐enriched fractions (p10, p100) as a percentage of total estimated expression (p10 + p100+S100 by C q) in ascending order, from miR‐27a‐3p (7.9%) to miR‐28‐3p (32.0%). (C) miRNAs in each fraction (10 000 g pellet = p10, 110 000 g pellet = p100, 110 000 g supernatant = S100, and) are significantly correlated (P < 0.0001, Spearman).
qPCR Validation
Individual stem‐loop RT/hydrolysis probe qPCR assays were used to verify TLDA results for eleven selected miRNAs plus miR‐375‐3p (not included on the array), which was also measured because of a reported association with goblet cells [54]. Some miRNAs were chosen due to high expression levels. miR‐181a‐5p was measured due to its association with endometrial cells [55, 56]. miR‐125b‐5p has been reported as a diagnostic marker of endometriosis [57]. Other miRNAs (miRs‐186‐5p, ‐451a‐5p, ‐200c‐3p, ‐222‐3p, ‐193b‐3p) were selected based on our previous experience and results from other studies evaluating miRNAs in the context of HIV‐1 and SIV infections. Results of qPCR assays, adjusted by miR‐16‐5p for each sample (since we found relatively low qPCR variation of miR‐16‐5p, a commonly used normalizer [58]), are shown in Fig. 3A. Figure 3B compares miRNA ranks (1–11) by TLDA and individual qPCR, which are generally in concordance. Note that expression of RBC miRNA miR‐451a‐5p was low, suggesting minimal contamination from blood for most samples.
Fig. 3.
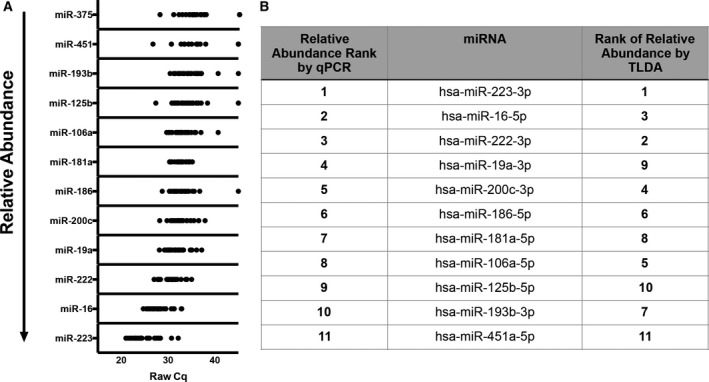
miRNA qPCR validation. (A) qPCR validation for UC pellet samples, all subjects (n = 6), and time points (individual dots). (B) Ranks of abundant miRNAs by qPCR and TLDA.
miRNA association with retroviral infection status
An association of miRNA abundance with infection status could yield novel biomarkers as well as clues to roles of miRNA in modulating infection. However, the small number of subjects in our study was a challenge. Nevertheless, by considering all subjects and time points together for both infected and uninfected subjects, microarray data suggested a slightly reduced abundance of miRs‐186‐5p, ‐222‐3p, and ‐200c‐3p in infected samples (Fig. 4A) based on statistical analysis of ∆C q values, while qPCR revealed differential abundance of miRs‐186‐5p and ‐125b‐5p (Fig. 4B). miR‐186‐5p was thus identified by both techniques as potentially associated with retroviral infection.
Fig. 4.
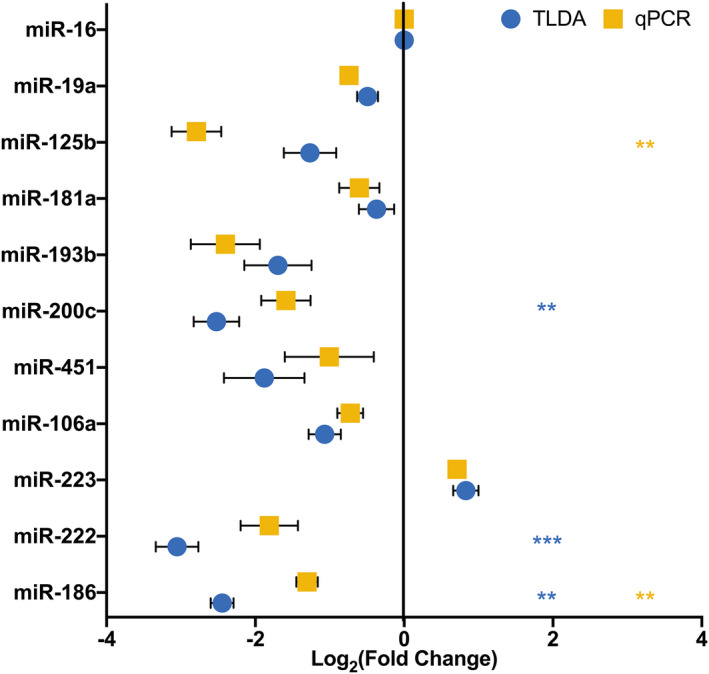
miR‐186‐5p downregulation: SIV. miR‐186‐5p fold change for all infected animals (n = 4) was determined using ∆∆C t method using miR‐16 and uninfected animals (n = 2) as controls. Log2 (fold change) for both TLDA and qPCR analyses was plotted for 11 selected validation miRNAs. Statistical analyses were performed on ∆C t values. For TLDA, miRs‐186, ‐222, and ‐200c were significantly less abundant in the CVL p100 fraction of infected subjects (mean ± SEM, multiple t‐test, Bonferroni–Dunn correction), **P < 0.01, ***P < 0.001. For qPCR analysis, miRs‐186 and ‐125b were significantly less abundant (multiple t‐test, Bonferroni–Dunn correction), **P < 0.01, ***P < 0.001.
miR‐186‐5p transfection has minimal effects on cellular HIV RNA abundance but reduces p24 release from monocyte‐derived macrophages
To assess a possible influence of miR‐186‐5p (“miR‐186”) on retroviral replication, we introduced double‐stranded miR‐186‐5p mimic or control RNA into monocyte‐derived macrophages derived from three donors 24 h before infecting the cells or not with HIV. Upon M‐CSF stimulation, the primary macrophages should be activated toward the alternative M2 phenotype [59, 60, 61]. Post‐infection, EVs and miRNAs were mostly likely released from uninfected and infected macrophages alike. At days 3 and 6 postinfection, we quantitated full‐length HIV‐1 transcript using a gag qPCR with standard curve. In cells from only one of three donors were fewer HIV‐1 copies associated with miR‐186‐5p mimic transfection (Fig. 5). Overall, there was no statistically significant difference in HIV RNA between the conditions.
Fig. 5.
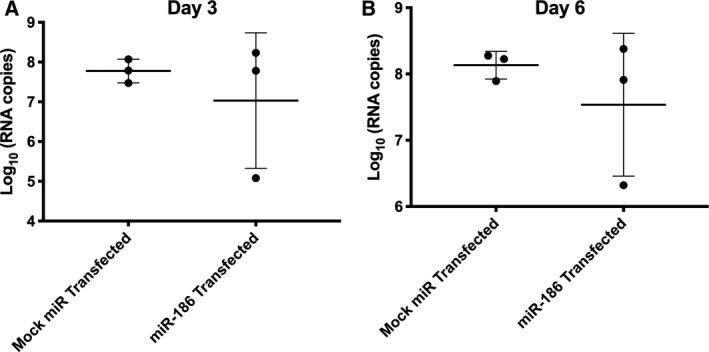
miRNA‐186‐5p mimic transfection inconsistently suppresses HIV‐1 gag mRNA production. Apparent downregulation of gag mRNA (qPCR assay with standard curve) was observed in miR‐186‐transfected monocyte‐derived macrophages from only one of three donors compared with mock miRNA‐transfected cells. Overall, results were insignificant by t‐test (mean ± SEM), P > 0.1, with multiple replicates of cells from human donors (n = 3).
However, at the same time points and also out to 9 days postinfection, a different result was seen for capsid p24 release into the supernatant. For infected but untransfected cells, measurable p24 was observed by 3 dpi, and p24 counts increased by twofold or more by 9 dpi (Fig. 6A) for multiple replicate experiments with cells from three donors. Compared with infected, untreated controls, mock‐transfected cells (not shown), and cells transfected with a negative control RNA (labeled with a fluorophore to assess transfection efficiency), miR‐186‐5p transfection was associated with a significant decline of released p24 at all time points (ANOVA with Bonferroni correction; Fig. 6B–D). The negative control condition showed a suppressive trend that reached nominal significance at 9 dpi. However, miR‐186‐associated suppression was significantly greater at all time points.
Fig. 6.
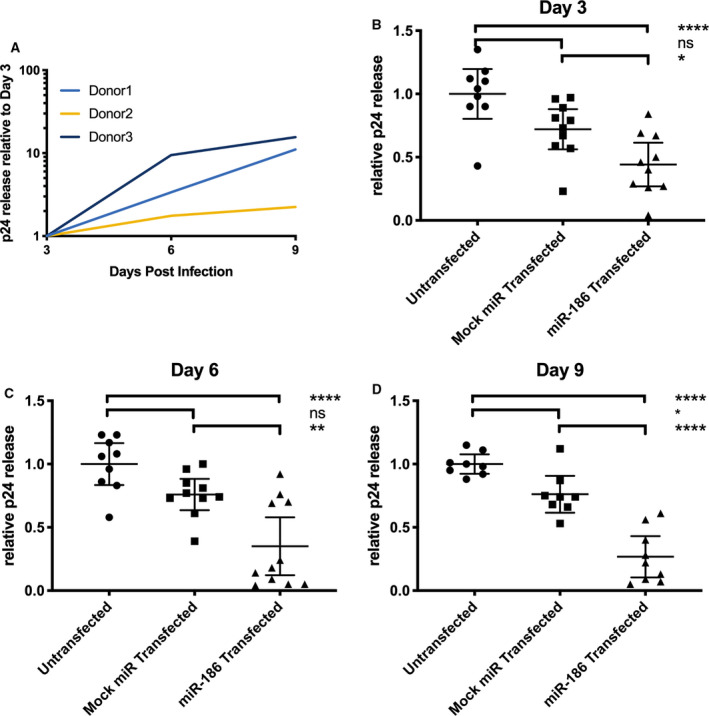
miRNA‐186‐5p inhibits p24 release. Monocyte‐derived macrophages from human donors were infected with HIV‐1 BaL. (A) p24 production increased > 2‐fold for all donors from 3 to 9 days postinfection (dpi), untreated cells. (B–D) Transfection of miR‐186‐5p mimic was associated with a decrease of p24 release compared with untransfected controls and mock miRNA mimic‐transfected controls at the indicated time points; ns = not significant, *P < 0.05, **P < 0.01, ****P < 0.0001 (mean ± SEM, ANOVA followed by Bonferroni correction for multiple tests). Results were from a total of eight to 11 replicate experiments with cells from all human donors (n = 3).
p24 inhibition by miR‐186‐5p is correlated with transfection efficiency
Despite the statistical significance of miR‐186‐5p‐associated p24 inhibition, substantial variability was observed, including between donors/experiments; we therefore hypothesized that either donor‐ or experiment‐specific factors were responsible for the variability. The transfection experiments were repeated using macrophages from five additional donors (labeled 1–5). While significant but variable inhibition of p24 release after miR‐186‐5p transfection was observed for three donors (1, 2, and 5), little or no inhibition was seen for donors 3 and 4 (Fig. 7). It should be noted that miR‐186‐5p antisense inhibitors were also introduced in these experiments. While they did not significantly increase HIV p24 release (Fig. 7), they also did not achieve a consistent knockdown of native miR‐186‐5p (Fig. 8A).
Fig. 7.
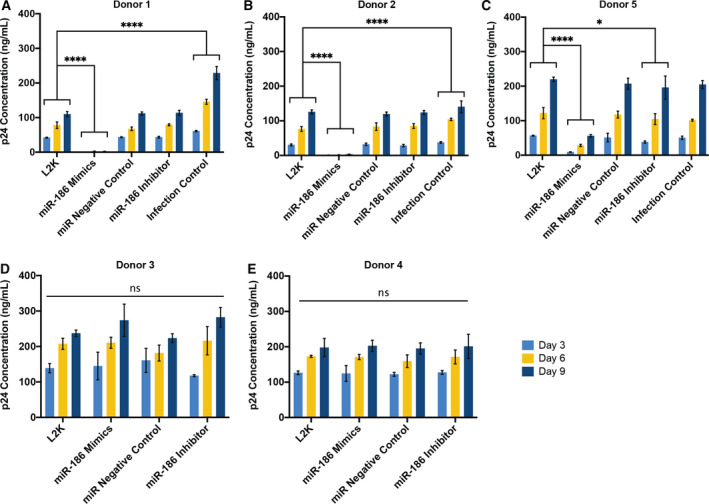
miRNA‐186‐5p inhibits p24 release in a donor‐specific manner. Monocyte‐derived macrophages from human donors were infected with HIV‐1 BaL. (A–C) Compared with mock‐transfected controls, transfection of miR‐186 mimic was associated with a significant decrease of p24 production from 3 to 9 days postinfection, respectively (dpi) in donors 1, 2, and 5 (biological replicate n = 3, technical replicate n = 2). (D, E) For donors 3 and 4, transfection of miR‐186 mimic was ineffective in inhibiting p24 release compared with mock‐transfected controls (biological replicate n = 3, technical replicate n = 2); ns = not significant, *P < 0.05, ****P < 0.0001 (mean ± SEM, two‐way ANOVA followed by Bonferroni correction for multiple tests).
Fig. 8.
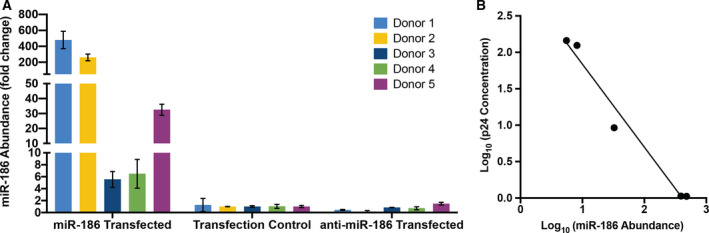
miR‐186‐5p abundance post‐transfection and correlation with p24 release. (A) Abundance of miR‐186‐5p in macrophages post‐transfection, as assessed by qPCR and compared (fold change) with the average of control macrophages (Mean ± SD). (B) Correlation of macrophage miR‐186‐5p and p24 concentration released in supernatant 3 days postinfection. P(two‐tailed) = 0.0019 (correlation), R 2 = 0.9731 (linear regression).
One experimental variable that could affect the degree of inhibition is the efficiency with which the miRNA mimic is delivered into the cells. Since this variable was not assessed in our previous experiments, we measured it for the five new experiments. Despite using the same nominal concentrations of miRNA mimics for our experiments, a nearly 100‐fold range of miR‐186‐5p concentration was observed between the lowest and highest efficiency transfections (Fig. 8A), which increased miR‐186‐5p levels from around 5‐fold to nearly 500‐fold, respectively. Strikingly, the miR‐186‐5p level was inversely correlated with released p24 across these five donors (Fig. 8B).
Discussion
Cervicovaginal lavage EVs and exRNPs, like EVs in the uterus [62, 63], may offer information about the health of the reproductive tract and may also facilitate or block transmission of infectious agents. Proteomic analyses of human [64] and rhesus macaque [65] CVL have suggested a core proteome and a highly variable proteome that responds to changes in pregnancy status, menstruation, infection, and other stressors. However, exRNA and EV profiles are less understood in this compartment. Thus, one major finding of this study is a partial profile of miRNAs of EV‐enriched and EV‐depleted fractions of CVL fluid of primates. We report that EVs can be liberated from vaginal secretions by lavage and that these EVs can be concentrated using a standard stepped centrifugation procedure, with enrichment of positive (membrane‐associated) markers while a cellular negative control was not detected.
Both EV‐replete and EV‐depleted fractions of CVL contained abundant miRNA. As reported for other biological fluids [37, 38], miRNA concentration was highest in the EV‐depleted CVL fractions, not in EV‐enriched UC pellets, consistent with packaging of most extracellular miRNA into exRNPs; the function, if any, of extracellular miRNAs in the cervicovaginal tract of healthy individuals remains to be determined. We observed minimal differences in extracellular miRNA profiles between SIV‐infected and uninfected subjects or, surprisingly, even during the menstrual cycle, suggesting a certain stability of extracellular miRNA in the compartment. Correlation of miRNA concentrations in EV‐depleted and EV‐replete fractions was also apparent. Based on relative abundance compared with miRNAs of other cellular/tissue origins (e.g., heart‐ and lung‐specific miR‐126, kidney‐specific miR‐196b, and liver‐specific miR‐192) [66, 67], miRNAs in EVs and exRNPs of CVL are likely derived from epithelial cells (including goblet cells) and cells of the immune system (as suggested, e.g., by myeloid‐enriched miR‐223 and lymphocyte‐enriched miR‐150) [68]. Of the most abundant miRNAs we identified, some have been ascribed tumor‐suppressive roles in cancers [69, 70, 71, 72, 73, 74, 75]. Also, miR‐223 and miR‐150 have been described as “anti‐HIV” miRNAs [76] among a variety of reported antiretroviral sRNAs, both host and viral [77, 78, 79, 80, 81, 82]. Given their relative abundance in the vaginal tract, a common site for HIV infection, these miRNAs may contribute to antiviral defenses.
Along these lines, a second major finding of this study is a possible role for miR‐186‐5p in antiretroviral defense, bolstered by the observation that exogenous miR‐186‐5p transfection efficiency correlates inversely with HIV p24 release. Previous publications have identified protein constituents in the CVL with anti‐HIV efficacies (e.g., [7, 21, 22]). Our identification of miRNA as a potential anti‐HIV agent adds an element of complexity to the picture of tissue‐specific antiretroviral defense. In contrast with an early report of direct binding of host miRNAs to retroviral transcripts and subsequent suppression [76], it now appears that this mechanism of suppression may be relatively uncommon [83]. Anti‐HIV miRNAs may be more likely to exert effects through control of host genes instead (e.g., [84]). Our data also support the conclusion that reduction of HIV RNA levels is not the main mechanism for miR‐186‐mediated suppression of HIV release.
How, then, might miR‐186‐5p, whether endogenous or exogenous (therapeutically introduced) contribute to antiretroviral effects? Combining several miRNA target prediction, validation, and enrichment analysis approaches [85, 86, 87, 88, 89, 90, 91], we noticed a few putative miR‐186‐5p targets and related pathways that may merit follow‐up. One target of miR‐186‐5p that was validated experimentally by multiple methods is FOXO1 [92], an important contributor to apoptosis but also immunoregulation via IFNγ pathways. Another prominent validated target, P2X7R [93], is involved in membrane budding, T‐cell‐mediated cytotoxicity, cellular response to extracellular stimuli, and T‐cell homeostasis/proliferation. There is also evidence that miR‐186‐5p targets the HIV coreceptor CXCR4 [94]. Pathway enrichment analyses [90, 91] suggest that miR‐186‐5p targets participate significantly in infection‐related networks, including prion diseases, viral carcinogenesis, and responses to measles and herpes simplex virus infections. Although miRNA target prediction algorithms are imperfect, and validation efforts are of varying quality [95, 96], these findings may shed some light on how miR‐186‐5p is involved in responses to HIV.
We would like to emphasize several aspects of the study that open the door to future research:
We used stepped ultracentrifugation without density gradients because of the small sample volumes available. Although stepped ultracentrifugation remains a widely used method for EV enrichment [49, 97], subsequent gradients or alternative isolation methods could be attempted with larger volume samples to increase purity in future. Possibly, our study overestimates the abundance of miRNAs in CVL EVs, and differential packaging into EVs and exRNPs is masked by contamination of our EV preps with exRNPs.
Our qPCR array approach and focus on miRNAs leaves room for additional work. While we are confident that our array captured most of the abundant miRNAs in CVL, sequencing short and longer RNAs could reveal additional markers.
The small number of subjects and the absence of obvious menstrual cycle in infected subjects preclude strong conclusions about EV or miRNA associations with either infection or the menstrual cycle. For example, we did not observe the expected increase in miR‐451a or other RBC‐specific miRNAs during menstruation. However, since only two animals showed evidence of cycling, experiments with more subjects and larger sample volumes are needed.
Our previous criticisms of miRNA functional studies [98] also apply to our results here. Additional work is needed to assess the potential of miR‐186‐5p to regulate retrovirus production at endogenous levels, for example by showing that it is present in active RNPs [99] and that it interacts directly with specific host or viral targets. However, it is also important to note that miR‐186‐5p could have therapeutic benefit even if it must be delivered at supraphysiologic concentrations. Finally, it is possible, but must be demonstrated, that miR‐186‐5p acts in a paracrine fashion via EV or exRNP shuttles.
We have investigated the effects of miR‐186‐5p only in monocyte‐derived macrophages. We chose to begin with this cell type because of the abundance of miR‐223 and the known role of macrophages in the epithelium. We would like to reiterate the importance of other cell types in the vaginal tissue during HIV‐1 infection [100, 101]; thus, this antiviral effect of miR‐186‐5p should also be investigated in other cell types.
Overall, the results presented here support further development of CVL and its constituents as a window into the health of the cervicovaginal compartment in retroviral infection and beyond. Furthermore, delivery of miR‐186‐5p could act to suppress retrovirus release.
Conflicts of interest
The authors have no competing interests to declare. The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of the data; in the writing of the report; or in the decision to submit the report for publication.
Author contributions
ZZ, DCM, GVH, KAMP, and KWW involved in conceptualization; ZZ and KWW involved in data curation; ZZ, DCM, and KWW made formal analysis; KWW acquired funding; ZZ, DCM, KM, ZL, and BHP involved in the investigation; ZZ, KM, GVH, KAMP, and KWW contributed to methodology; KWW administrated the project; KAMP and KWW facilitated the resources; ZL and KWW supervised the study; ZZ, DCM, and KWW involved in visualization; KWW wrote the original draft; ZZ, DCM, and KWW Writing – review and editing.
Supporting information
Fig. S1. Specimen collection and sample processing workflow.
Fig. S2. miRNA profile of CVL fractions.
Fig. S3. miRNA‐186‐5p suppresses HIV‐1 gag mRNA production on Day 6 and inhibits p24 release on Day 3–6.
Table S1. Recovered volumes: CVL.
Table S2. NTA dilution factors, CVL.
Acknowledgements
The authors thank Robert Adams, Lauren Ostrenga, and Sarah Beck for contributions to these studies. The authors gratefully acknowledge the Oregon National Primate Research Center and David Erikson for hormone analyses and endocrinology advice and thank Barbara Smith of the JHU IBBS Microscope Facility for expert assistance with electron microscopy. Amanda Steele provided paid assistance with editing and organizing an early version of the manuscript. This research was funded by the Johns Hopkins University Center for AIDS Research, an NIH funded program, grant number P30AI094189 (pilot grant to KWW; ZZ and GVH were Baltimore HIV/AIDS scholars); by the US National Institutes of Health, DA040385, DA047807, AI144997, and MH118164 (to KWW); by UG3CA241694, supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director; and by the National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) and the National Institutes of Health, grant number P40 OD013117. DM and KM received support through NIH grant number T32 OD011089.
Data accessibility
Array data have been deposited with the Gene Expression Omnibus [102] as GSE107856. Data in other formats are available upon request. To the extent that sample quantities would allow, the minimal information for studies of extracellular vesicles recommendations for EV studies were followed [24], and the EV experiments have been registered with the EV‐TRACK knowledgebase [103] with preliminary EV‐TRACK code XL5296IL.
References
- 1. Hanson EK and Ballantyne J (2013) Highly specific mRNA biomarkers for the identification of vaginal secretions in sexual assault investigations. Sci Justice 53, 14–22. [DOI] [PubMed] [Google Scholar]
- 2. Jakubowska J, MacIejewska A, Pawłowski R and Bielawski KP (2013) MRNA profiling for vaginal fluid and menstrual blood identification. Forensic Sci Int Genet 7, 272–278. [DOI] [PubMed] [Google Scholar]
- 3. Park JL, Kwon OH, Kim JH, Yoo HS, Lee HC, Woo KM, Kim SY, Lee SH and Kim YS (2014) Identification of body fluid‐specific DNA methylation markers for use in forensic science. Forensic Sci Int Genet 13, 147–153. [DOI] [PubMed] [Google Scholar]
- 4. Hanson EK, Lubenow H and Ballantyne J (2009) Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem 387, 303–314. [DOI] [PubMed] [Google Scholar]
- 5. Liu J, Sun H, Wang X, Yu Q, Li S, Yu X and Gong W (2014) Increased exosomal microRNA‐21 and MicroRNA‐146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int J Mol Sci Int J Mol Sci 15, 758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Ostade X, Dom M, Tjalma W and Van Raemdonck G (2018) Candidate biomarkers in the cervical vaginal fluid for the (self‐)diagnosis of cervical precancer. Arch Gynecol Obstet 297, 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Raemdonck GAA, Tjalma WAA, Coen EP, Depuydt CE and Van Ostade XWM (2014) Identification of protein biomarkers for cervical cancer using human cervicovaginal fluid. PLoS One 9, e106488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gravett MG, Thomas A, Schneider KA, Reddy AP, Dasari S, Jacob T, Lu X, Rodland M, Pereira L, Sadowsky DW et al (2007) Proteomic analysis of cervical−vaginal fluid: identification of novel biomarkers for detection of intra‐amniotic infection. J Proteome Res 6, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Datcu R, Gesink D, Mulvad G, Montgomery‐Andersen R, Rink E, Koch A, Ahrens P and Jensena JS (2014) Bacterial vaginosis diagnosed by analysis of first‐void‐urine specimens. J Clin Microbiol 52, 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srinivasan S, Morgan MT, Fiedler TL, Djukovic D, Hoffman NG, Raftery D, Marrazzo JM and Fredricks DN (2015) Metabolic signatures of bacterial vaginosis supplementary file: Figure S1. MBio 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zevin AS, Xie IY, Birse K, Arnold K, Romas L, Westmacott G, Novak RM, McCorrister S, McKinnon LR, Cohen CR et al (2016) Microbiome composition and function drives wound‐healing impairment in the female genital tract. PLoS Pathog 12, e1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boggiano C and Littman DR (2007) HIV's vagina travelogue. Immunity 26, 145–147. [DOI] [PubMed] [Google Scholar]
- 13. Patel MV, Ghosh M, Fahey JV, Ochsenbauer C, Rossoll RM and Wira CR (2014) Innate immunity in the vagina (Part II): anti‐HIV activity and antiviral content of human vaginal secretions. Am J Reprod Immunol 72, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benki S, Mostad SB, Richardson BA, Mandaliya K, Kreiss JK and Overbaugh J (2008) Increased levels of HIV‐1‐infected cells in endocervical secretions after the luteinizing hormone surge. J Acquir Immune Defic Syndr 47, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zara F, Nappi RE, Brerra R, Migliavacca R, Maserati R and Spinillo A (2004) Markers of local immunity in cervico‐vaginal secretions of HIV infected women: implications for HIV shedding. Sex Transm Infect 80, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gardella B, Roccio M, Maccabruni A, Mariani B, Panzeri L, Zara F and Spinillo A (2011) HIV shedding in cervico‐vaginal secretions in pregnant women. Curr HIV Res 9, 313–320. [DOI] [PubMed] [Google Scholar]
- 17. Seaton KE, Ballweber L, Lan A, Donathan M, Hughes S, Vojtech L, Moody MA, Liao HX, Haynes BF, Galloway CG et al (2014) HIV‐1 specific IgA detected in vaginal secretions of HIV uninfected women participating in a microbicide trial in Southern Africa are primarily directed toward gp120 and gp140 specificities. PLoS One 9, e101863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghosh M, Fahey JV, Shen Z, Lahey T, Cu‐Uvin S, Wu Z, Mayer K, Wright PF, Kappes JC, Ochsenbauer C et al (2010) Anti‐HIV activity in cervical‐vaginal secretions from HIV‐positive and ‐negative women correlate with innate antimicrobial levels and IgG antibodies. PLoS One 5, e11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clemetson DB, Moss GB, Willerford DM, Hensel M, Emonyi W, Holmes KK, Plummer F, Ndinya‐Achola J, Roberts PL, Hillier S et al (1993) Detection of HIV DNA in cervical and vaginal secretions. Prevalence and correlates among women in Nairobi, Kenya. JAMA 269, 2860–2864. [PubMed] [Google Scholar]
- 20. Burgener A, Rahman S, Ahmad R, Lajoie J, Ramdahin S, Mesa C, Brunet S, Wachihi C, Kimani J, Fowke K et al (2011) Comprehensive proteomic study identifies serpin and cystatin antiproteases as novel correlates of HIV‐1 resistance in the cervicovaginal mucosa of female sex workers. J Proteome Res 10, 5139–5149. [DOI] [PubMed] [Google Scholar]
- 21. Drannik AG, Nag K, Yao XD, Henrick BM, Ball TB, Plummer FA, Wachihi C, Kimani J and Rosenthal KL (2012) Anti‐HIV‐1 activity of elafin depends on its nuclear localization and altered innate immune activation in female genital epithelial cells. PLoS One 7, e52738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgener A, Boutilier J, Wachihi C, Kimani J, Carpenter M, Westmacott G, Cheng K, Ball TB and Plummer F (2008) Identification of differentially expressed proteins in the cervical mucosa of HIV‐1‐resistant sex workers. J Proteome Res 7, 4446–4454. [DOI] [PubMed] [Google Scholar]
- 23. Yáñez‐Mó M, Siljander PR‐M, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J et al (2015) Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Witwer KW and Théry C (2019) Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J Extracell Vesicles 8, 1648167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meehan B, Rak J and Di Vizio D (2016) Oncosomes ‐ large and small: what are they, where they came from? J Extracell Vesicles 5, 33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. György B, Hung ME, Breakefield XO and Leonard JN (2015) Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol 55, 439–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte‐'t Hoen EN, Piper MG, Sivaraman S, Skog J et al(2013) Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 2, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith JA and Daniel R (2016) Human vaginal fluid contains exosomes that have an inhibitory effect on an early step of the HIV‐1 life cycle. AIDS 30: 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muth DC, McAlexander MA, Ostrenga LJ, Pate NM, Izzi JM, Adams RJ, Pate KAM, Beck SE, Karim BO and Witwer KW (2015) Potential role of cervicovaginal extracellular particles in diagnosis of endometriosis. Bmc Vet Res 11, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sergeeva AM, Pinzon Restrepo N and Seitz H (2013) Quantitative aspects of RNA silencing in metazoans. Biochem 78, 613–626. [DOI] [PubMed] [Google Scholar]
- 32. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ and Lötvall JO (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- 34. Aliotta JM, Sanchez‐Guijo FM, Dooner GJ, Johnson KW, Dooner MS, Greer KA, Greer D, Pimentel J, Kolankiewicz LM, Puente N et al (2007) Alteration of marrow cell gene expression, protein production, and engraftment into lung by lung‐derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells 25, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baj‐Krzyworzeka M, Szatanek R, Węglarczyk K, Baran J, Urbanowicz B, Brański P, Ratajczak MZ and Zembala M (2006) Tumour‐derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother 55, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FWN, Das S et al (2017) Obstacles and opportunities in the functional analysis of extracellular vesicle RNA ‐ an ISEV position paper. J Extracell Vesicles 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turchinovich A, Weiz L, Langheinz A and Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39, 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova‐Agadjanyan EL, Stirewalt DL et al (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Witwer KW, Sarbanes SL, Liu J and Clements JE (2011) A plasma microRNA signature of acute lentiviral infection: biomarkers of CNS disease. AIDS 204, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA and Kayser M (2010) MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT‐PCR confirmation. Int J Leg Med 124, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seashols‐Williams S, Lewis C, Calloway C, Peace N, Harrison A, Hayes‐Nash C, Fleming S, Wu Q and Zehner ZE (2016) High‐throughput miRNA sequencing and identification of biomarkers for forensically relevant biological fluids. Electrophoresis 37, 2780–2788. [DOI] [PubMed] [Google Scholar]
- 42. Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran‐Walters S, Dandekar S, Clapham PR, Smythies LE et al (2009) Macrophages in vaginal but not intestinal mucosa are monocyte‐like and permissive to human immunodeficiency virus type 1 infection. J Virol 83, 3258–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iijima N, Thompson JM and Iwasaki A (2008) Dendritic cells and macrophages in the genitourinary tract. Mucosal Immunol 1, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hill JA and Anderson DJ (1992) Human vaginal leukocytes and the effects of vaginal fluid on lymphocyte and macrophage defense functions. Am J Obstet Gynecol 166, 720–726. [DOI] [PubMed] [Google Scholar]
- 45. Greenhead P, Hayes P, Watts PS, Laing KG, Griffin GE and Shattock RJ (2000) Parameters of human immunodeficiency virus infection of human cervical tissue and inhibition by vaginal virucides. J Virol 74, 5577–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen R, Richter HE and Smith PD (2011) Early HIV‐1 target cells in human vaginal and ectocervical mucosa. Am J Reprod Immunol 65, 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hladik F, Lentz G, Akridge RE, Peterson G, Kelley H, McElroy A and McElrath MJ (1999) Dendritic cell–T‐cell interactions support coreceptor‐independent human immunodeficiency virus type 1 transmission in the human genital tract. J Virol 73, 5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rahman S, Rabbani R, Wachihi C, Kimani J, Plummer FA, Ball TB and Burgener A (2013) Mucosal serpin A1 and A3 levels in HIV highly exposed sero‐negative women are affected by the menstrual cycle and hormonal contraceptives but are independent of epidemiological confounders. Am J Reprod Immunol 69, 64–72. [DOI] [PubMed] [Google Scholar]
- 49. Thery C, Amigorena S, Raposo G and Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 50. McAlexander MA, Phillips MJ and Witwer KW (2013) Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet 4, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR et al (2005) Real‐time quantification of microRNAs by stem‐loop RT‐PCR. Nucleic Acids Res 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Witwer KW, Sarbanes SL, Liu J and Clements JE (2011) A plasma microRNA signature of acute lentiviral infection: biomarkers of central nervous system disease. AIDS 25, 2057–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A and Speleman F (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, research 0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo‐Russo S, Avnit‐Sagi T, Cojocaru G, Zreik F et al (2011) Epithelial microRNAs regulate gut mucosal immunity via epithelium–T cell crosstalk. Nat Immunol 12, 239–246. [DOI] [PubMed] [Google Scholar]
- 55. Jurcevic S, Olsson B and Klinga‐Levan K (2014) MicroRNA expression in human endometrial adenocarcinoma. Cancer Cell Int 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jayaraman M, Radhakrishnan R, Mathews CA, Yan M, Husain S, Moxley KM, Song YS and Dhanasekaran DN (2017) Identification of novel diagnostic and prognostic miRNA signatures in endometrial cancer. Genes Cancer 8, 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cosar E, Mamillapalli R, Ersoy GS, Cho SY, Seifer B and Taylor HS (2016) Serum microRNAs as diagnostic markers of endometriosis: a comprehensive array‐based analysis. Fertil Steril 106, 402–409. [DOI] [PubMed] [Google Scholar]
- 58. Schwarzenbach H, da Silva AM, Calin G and Pantel K (2015) Data normalization strategies for MicroRNA quantification. Clin Chem 61, 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verreck FAW, De Boer T, Langenberg DML, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, De Waal‐Malefyt R and Ottenhoff THM (2004) Human IL‐23‐producing type 1 macrophages promote but IL‐10‐producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA 101, 4560–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Martinez FO, Gordon S, Locati M and Mantovani A (2006) Transcriptional profiling of the human monocyte‐to‐macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177, 7303–7311. [DOI] [PubMed] [Google Scholar]
- 61. Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD and Hamilton JA (2012) Defining GM‐CSF– and macrophage‐CSF–dependent macrophage responses by in vitro models. J Immunol 188, 5752–5765. [DOI] [PubMed] [Google Scholar]
- 62. Nguyen HPT, Simpson RJ, Salamonsen LA and Greening DW (2016) Extracellular vesicles in the intrauterine environment: challenges and potential functions. Biol Reprod 95, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Campoy I, Lanau L, Altadill T, Sequeiros T, Cabrera S, Cubo‐Abert M, Pérez‐Benavente A, Garcia A, Borrós S, Santamaria A et al (2016) Exosome‐like vesicles in uterine aspirates: a comparison of ultracentrifugation‐based isolation protocols. J Transl Med 14, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zegels G, Aa G, Raemdonck V, Coen EP, Tjalma WA, Wm X and Ostade V (2009) Comprehensive proteomic analysis of human cervical‐vaginal fluid using colposcopy samples. Proteome Sci 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gravett MG, Thomas A, Schneider KA, Reddy AP, Dasari S, Jacob T, Lu X, Rodland M, Pereira L, Sadowsky DW et al (2007) Proteomic analysis of cervical‐vaginal fluid: identification of novel biomarkers for detection of intra‐amniotic infection. J Proteome Res 6, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guo Z, Maki M, Ding R, Yang Y, Zhang B and Xiong L (2014) Genome‐wide survey of tissue‐specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Panwar B, Omenn GS and Guan Y (2017) miRmine: a database of human miRNA expression profiles. Bioinformatics 33, btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF and Tewari M (2012) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res 5, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luo P, Wang Q, Ye Y, Zhang J, Lu D, Cheng L, Zhou H, Xie M and Wang B (2019) MiR‐223‐3p functions as a tumor suppressor in lung squamous cell carcinoma by miR‐223‐3p‐mutant p53 regulatory feedback loop. J Exp Clin Cancer Res 38, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ et al (2018) miR‐223‐3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther 26, 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lawrence MJY (2015) miR‐203 functions as a tumor suppressor by inhibiting epithelial to mesenchymal transition in ovarian cancer. J Cancer Sci Ther 7, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin Q, Zhou L and Sun X (2016) MiRNA‐203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep 6, 28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang S, Zhang R, Claret FX and Yang H (2014) Involvement of microRNA‐24 and DNA methylation in resistance of nasopharyngeal carcinoma to ionizing radiation. Mol Cancer Ther 13, 3163–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fang ZH, Wang SL, Zhao JT, Lin ZJ, Chen LY, Su R, Xie ST, Carter BZ and Xu B (2016) MIR‐150 exerts antileukemia activity in vitro and in vivo through regulating genes in multiple pathways. Cell Death Dis 7, e2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ito M, Teshima K, Ikeda S, Kitadate A, Watanabe A, Nara M, Yamashita J, Ohshima K, Sawada K and Tagawa H (2014) MicroRNA‐150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T‐cell lymphoma. Blood 123, 1499–1511. [DOI] [PubMed] [Google Scholar]
- 76. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G and Zhang H (2007) Cellular microRNAs contribute to HIV‐1 latency in resting primary CD4+ T lymphocytes. Nat Med 13, 1241–1247. [DOI] [PubMed] [Google Scholar]
- 77. Swaminathan S, Murray DD and Kelleher AD (2013) miRNAs and HIV: unforeseen determinants of host‐pathogen interaction. Immunol Rev 254, 265–280. [DOI] [PubMed] [Google Scholar]
- 78. Sisk JM, Witwer KW, Tarwater PM and Clements JE (2013) SIV replication is directly downregulated by four antiviral miRNAs. Retrovirology 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang X, Ye L, Zhou Y, Liu MQ, Zhou DJ and Ho WZ (2011) Inhibition of anti‐HIV microRNA expression: a mechanism for opioid‐mediated enhancement of HIV infection of monocytes. Am J Pathol 178, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Mendez C, Gelgor L et al (2012) Differential regulation of the Let‐7 family of microRNAs in CD4+ T cells alters IL‐10 expression. J Immunol 188, 6238–6246. [DOI] [PubMed] [Google Scholar]
- 81. Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T and Kashanchi F (2007) HIV‐1 TAR element is processed by Dicer to yield a viral micro‐RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent‐Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O et al (2012) Microprocessor, Setx, Xrn2, and Rrp6 co‐operate to induce premature termination of transcription by RNAPII. Cell 150, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH and Cullen BR (2013) In‐depth analysis of the interaction of HIV‐1 with cellular microRNA biogenesis and effector mechanisms. MBio 4, e000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sung TL and Rice AP (2009) miR‐198 inhibits HIV‐1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog 5, e1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY et al (2014) miRTarBase update 2014: an information resource for experimentally validated miRNA‐target interactions. Nucleic Acids Res 42, D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM et al (2011) miRTarBase: a database curates experimentally validated microRNA‐target interactions. Nucleic Acids Res 39, D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T and Hatzigeorgiou AG (2011) TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res 40, D222–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D et al (2015) DIANA‐TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 43, D153–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lagana A, Forte S, Giudice A, Arena MR, Puglisi PL, Giugno R, Pulvirenti A, Shasha D and Ferro A (2009) miRo: a miRNA knowledge base. Database 2009, bap008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M and Hatzigeorgiou AG (2009) DIANA‐mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics 25, 1991–1993. [DOI] [PubMed] [Google Scholar]
- 91. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and Hatzigeorgiou AG (2015) DIANA‐miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43, W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Myatt SS, Wang J, Monteiro LJ, Christian M, Ho K‐K, Fusi L, Dina RE, Brosens JJ, Ghaem‐Maghami S and Lam EW‐F (2010) Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res 70, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhou L, Qi X, Potashkin JA, Abdul‐Karim FW and Gorodeski GI (2008) MicroRNAs miR‐186 and miR‐150 down‐regulate expression of the pro‐apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′‐untranslated region of the gene that decrease steady‐state levels of the transcript. J Biol Chem 283, 28274–28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Niinuma T, Kai M, Kitajima H, Yamamoto E, Harada T, Maruyama R, Nobuoka T, Nishida T, Kanda T, Hasegawa T et al (2017) Downregulation of miR‐186 is associated with metastatic recurrence of gastrointestinal stromal tumors. Oncol Lett 14, 5703–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Paraskevopoulou MD, Vlachos IS and Hatzigeorgiou AG (2016) DIANA‐TarBase and DIANA suite tools: studying experimentally supported microRNA targets. Curr Protoc Bioinformatics 55, 12.14.1–12.14.18. [DOI] [PubMed] [Google Scholar]
- 96. Ji Diana Lee Y, Kim V, Muth DC and Witwer KW (2015) Validated microRNA target databases: an evaluation. Drug Dev Res 76, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gardiner C, Di Vizio D, Sahoo S, Théry C, Witwer KW, Wauben M and Hill AF (2016) Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles 5, 32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Witwer KW and Halushka MK (2016) Towards the promise of microRNAs ‐ enhancing reproducibility and rigor in microRNA research. RNA Biol.13, 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. La Rocca G, Olejniczak SH, Gonzalez AJ, Briskin D, Vidigal JA, Spraggon L, DeMatteo RG, Radler MR, Lindsten T, Ventura A et al (2015) In vivo, argonaute‐bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci USA 112, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Carias AM, McCoombe S, McRaven M, Anderson M, Galloway N, Vandergrift N, Fought AJ, Lurain J, Duplantis M, Veazey RS et al (2013) Defining the interaction of HIV‐1 with the mucosal barriers of the female reproductive tract. J Virol 87, 11388–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miller CJ and Shattock RJ (2003) Target cells in vaginal HIV transmission. Microbes Infect 5, 59–67. [DOI] [PubMed] [Google Scholar]
- 102. Clough E and Barrett T (2016) The gene expression omnibus database. Methods Mol Biol 1418, 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Van Deun J, Mestdagh P, Agostinis P, Akay Ö, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier L et al (2017) EV‐TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods 14, 228–232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Specimen collection and sample processing workflow.
Fig. S2. miRNA profile of CVL fractions.
Fig. S3. miRNA‐186‐5p suppresses HIV‐1 gag mRNA production on Day 6 and inhibits p24 release on Day 3–6.
Table S1. Recovered volumes: CVL.
Table S2. NTA dilution factors, CVL.
Data Availability Statement
Array data have been deposited with the Gene Expression Omnibus [102] as GSE107856. Data in other formats are available upon request. To the extent that sample quantities would allow, the minimal information for studies of extracellular vesicles recommendations for EV studies were followed [24], and the EV experiments have been registered with the EV‐TRACK knowledgebase [103] with preliminary EV‐TRACK code XL5296IL.


