Figure 5.
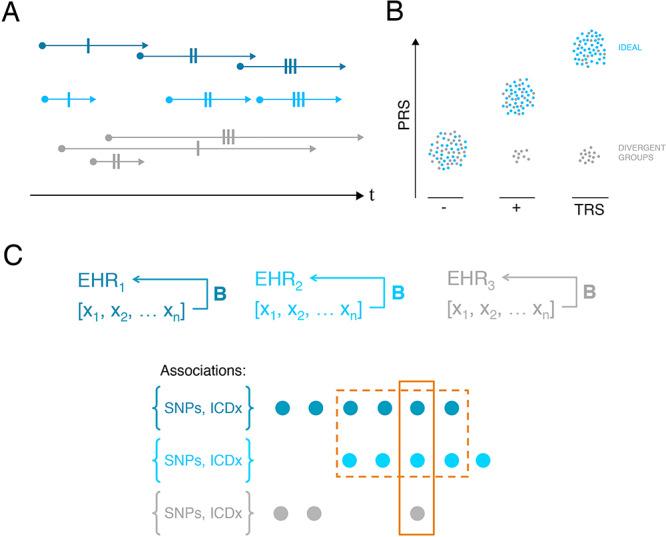
Approaches to detect biases in EHR. (A) Consider order and convergence of diagnoses across the lifespan. Here, we consider three individuals (dark blue, light blue and grey), with identical variables within the EHR (single-dashed lines represent identical diagnoses, etc.). From a retrospective, ‘snapshot’ perspective, all individuals appear identical; however, order and persistence of diagnoses clearly differ. (B) Leverage known biology. Here, we illustrate how polygenic risk scores might be applied to test accuracy of phenotype definitions. In this case, we consider controls (‘-’), schizophrenia cases (‘+’) and individuals with ‘TRS’. If these definitions are accurately assigned, we expect increased PRS in cases versus controls and in TRS versus others (blue). If however some bias affects phenotype assignment, we may identify a group of individuals with divergent PRS (grey). (C) Here, we illustrate three EHR (EHR1, EHR2 and EHR3), with overlapping but not identical sets of biases B and phenotypes [x1…xN]. Genotypic and phenotypic associations {SNPs, ICDx}, which are present across multiple different EHR are more likely to represent true biological signal rather than representing biased inferences.
