Figure 4.
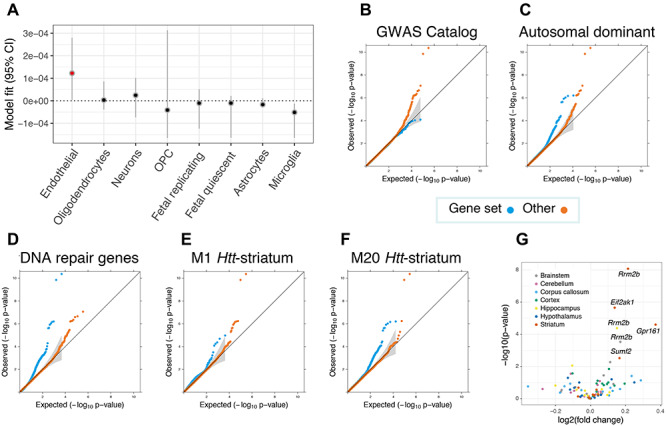
Enrichment analyses revealed brain cell types, striatal coexpression modules and gene sets that are most relevant for modifying the clinical onset of Huntington disease (HD). (A) Inferring relevant brain cell types from the GWAS data supplemented with cortical single cell sequencing information suggests that brain microvascular endothelial cells (annotated in red) could be relevant for the trait (P < 0.05). Cell types are ranked according to parameter estimates. Representative Q–Q plots for significantly enriched or depleted S-PrediXcan modifier associations for genes belonging to various gene sets (B–D), as well as coexpression networks that are related to murine Htt-CAG size over time (E and F). M20Htt-striatum genes are involved in p53 signaling and Brca1 in DNA damage response, whereas M1Htt-striatum genes are implicated in protein ubiquitination, tRNA charging and HD signaling. (G) Volcano plot for prioritized transcriptomic modifier genes in in Q175 mice versus wild type at 6 months of age for six brain-related tissues. Significantly dysregulated genes after correcting for multiple testing corrected are annotated, with all genes differentially expressed in the brain were up-regulated in mice with expanded Htt CAGs.
