Abstract
Frontotemporal dementia (FTD) is the second most prevalent form of pre-senile dementia after Alzheimer’s disease. Amyotrophic lateral sclerosis (ALS) can overlap genetically, pathologically and clinically with FTD indicating the two conditions are ends of a spectrum and may share common pathological mechanisms. FTD–ALS causing mutations are known to be involved in endosomal trafficking and RNA regulation. Using an unbiased genome-wide genetic screen to identify mutations affecting an FTD–ALS-related phenotype in Drosophila caused by CHMP2BIntron5 expression, we have uncovered repressors of retrovirus (RV) activity as modifiers of CHMP2BIntron5 toxicity. We report that neuronal expression of CHMP2BIntron5 causes an increase in the activity of the endogenous Drosophila RV, gypsy, in the nervous system. Genetically blocking Drosophila gypsy activation and pharmacologically inhibiting viral reverse transcriptase activity prevents degenerative phenotypes observed in fly and rat neurons. These findings directly link endosomal dysfunction to RV de-repression in an FTD–ALS model without TDP-43 pathology. These observations may contribute an understanding to previous discoveries of RV activation in ALS affected patients.
Keywords: retrovirus, frontotemporal dementia, amyotrophic lateral sclerosis, Drosophila, gypsy
Introduction
Frontotemporal dementia (FTD) is the second most common form of dementia in individuals under 60 years of age (1). FTD is characterized by atrophy of the frontal and temporal lobes. Within these brain regions, von Economo neurons are first affected triggering behavioral changes and cognitive impairment (2). FTD can co-occur with amyotrophic lateral sclerosis (ALS) and both conditions share clinical, neuropathological and genetic features, suggesting they may constitute two ends of a disease continuum (3–5) with approximately 15% of FTD patients developing ALS and vice versa (6).
Mutations in chromatin-modifying protein 2B (CHMP2B) have been found to cause rare cases of FTD (7,8), but have also been identified in some ALS cases (9–11) and in FTD–ALS (12). CHMP2B encodes a core component of the endosomal sorting complex required for transport-III (ESCRT-III) involved in transport of ubiquitinated proteins from the cell membrane to the lysosome (13). The pathological FTD-causing CHMP2BIntron5 mutation arises from a C-terminal truncation of the protein, perturbing endosomal trafficking resulting in autophagosome accumulation leading to neurodegeneration. The CHMP2BIntron5 mutation causes a form of FTD with no apparent TAR DNA-binding protein 43 (TDP-43), FUS or tau proteinopathy (14). Previous studies have established a Drosophila model of CHMP2BIntron5 (15), which has facilitated the unbiased screening for genetic modifiers of CHMP2BIntron5 toxicity identifying innate immune activation, autophagosomal and endosomal dysfunction (15–18). Here, we describe the identification of retrovirus (RV) reactivation as a potent modifier of the CHMP2BIntron5 phenotype.
Endogenous RVs are present in most eukaryotic genomes, accumulating in heterochromatic regions (19). They can act as genetically mobile elements that can replicate and move within the genome negatively affecting host health. RVs contain three open reading frames (ORFs) consisting of group-specific antigen (GAG), polymerase (POL) and envelope (ENV) components that upon activation, encode the capsid, reverse transcriptase and ENV protein of the virus, respectively. RVs can be initially repressed via cellular antiviral mechanisms and later domesticated and endogenized within the genome over time, with gain-and-loss of ORFs and ORFs evolving to generate novel proteins (20). In order to prevent RV activation, transposition and infection, eukaryotic cells have developed potent RV suppression mechanisms: small RNA molecules associated with the piwi protein termed piRNAs (21,22), which are known to act in the germline, and siRNAs that have a somatic role (23). An increase in RV expression has been documented in FTD–ALS postmortem tissue (24–26) suggesting that the RV silencing machinery might be disrupted potentially through TDP-43 dysfunction (27–29). An increase in the reverse transcriptase activity of human endogenous RV (hERV)-K has been documented in ALS patient serum and cerebrospinal fluid but the link between ALS pathology and RV abundance has yet to be fully identified (30). Neuronal expression of the hERV-K ENV protein in mice induced an ALS-like condition with loss of upper- and lower-motor neurons (26). These findings have been followed by clinical studies using nucleoside reverse transcriptase inhibitors as potential therapeutic treatments for ALS (31). Why RVs might be de-repressed in ALS tissue in some patients and if they may contribute to pathology has yet to be fully elucidated.
We have identified mutations in genes functioning in RV repression as dominant genetic enhancers and suppressors of CHMP2BIntron5 expression in Drosophila eye. Mutations, knockdown or overexpression of the RV repression component genes encoding Brother-of-Yb (BoYb), piwi, Sister-of-Yb (SoYb) and Vreteno (Vret) are seen to enhance and suppress an innate immune activation eye phenotype associated with CHMP2BIntron5 expression (15). Quantification of RV insertions in genomic DNA from flies neuronally expressing CHMP2BIntron5 shows an elevation of gypsy elements inserted in the genome with a concomitant increase in expression of the gypsy ENV protein. We show that knockdown of ENV protein via gypsy-RNAi in the presence of CHMP2BIntron5 alleviates photoreceptor cell death. Finally, we demonstrate that the addition of reverse transcriptase inhibitors in primary mammalian neurons transfected with CHMP2BIntron5 significantly rescues the neuronal dendritic retraction caused by CHMP2BIntron5 expression.
Results
A genetic screen in Drosophila identifies RV repression components as modifiers of CHMP2BIntron5 toxicity
The CHMP2BIntron5 mutation was identified in a large Danish family causing FTD on chromosome 3 [FTD-3; (7)]. In this study, we overexpressed this human mutant form in the Drosophila eye using the glass multimer reporter (GMR) promoter. This induces a rough eye with melanotic deposits [Fig. 1A; (15)]. The melanotic spots represent an innate immune activation and we principally quantify the level of melanization in our screen as a measure of FTD–ALS-related neuronal death and dysfunction (15). This model has identified several modifiers of CHMP2BIntron5 (15–18). From a genome-wide unbiased screen for enhancers and suppressors of this phenotype, we identified a heterozygous mutation in BoYb as a dominant genetic enhancer of CHMP2BIntron5 related toxicity (Fig. 1A and B). BoYb knockdown via RNAi potentiated the CHMP2BIntron5 melanization phenotype whereas overexpression reduced the melanization of the eye caused by CHMP2BIntron5 (Fig. 1A). BoYb is a component of the Yb-nuage piRNA regulatory machinery (32). To confirm and extend this data in vivo, a targeted screen using mutations and transgenes for other components of the RV repression machinery was performed to test for dominant modification of the CHMP2BIntron5 eye expression phenotype. Novel enhancers and suppressors identified included piwi, SoYb and Vret [Fig. 1A–D; (33)]. Enhancement of melanization by piwi was consistent for two additional identified piwi mutant alleles (Fig. 1C and D). Each of these are core components of the Drosophila Yb-nuage complex involved in RV repression (34). In control flies driving GMR-Gal4, these crosses had minimal effects on eye degeneration (Fig. 1A).
Figure 1.
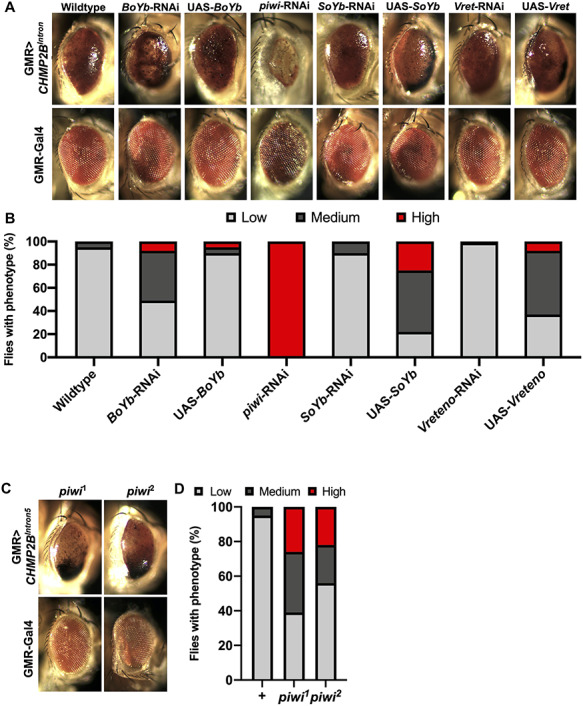
Manipulating levels of BoYb, piwi, SoYb and Vret perturbs melanization in the CHMP2BIntron5 expressing Drosophila eye. Representative eye of GMR-Gal4:UAS-CHMP2BIntron5/+ (GMR > CHMP2BIntron5) (A). BoYb knockdown via RNAi showed an enhancement on the eye blackening, whereas overexpression rescued the melanization caused by CHMP2BIntron5 expression. Reduction of piwi levels via Piwi-RNAi strongly increased the CHMP2BIntron5 eye melanization. A single copy of SoYb-RNAi and Vret-RNAi reduced the toxicity on the eye caused by CHMP2BIntron5, whereas SoYb and Vret overexpression enhanced the CHMP2BIntron5 melanization phenotype. (B) Quantification of the eye melanization severity from A genotypes (n = 100). (C) A single copy of the mutant alleles piwi1–2 enhanced the toxicity of the eye melanization caused by CHMP2BIntron5 expression. (D) Quantification of the eye melanization severity from C genotypes (n = 100).
Neuronal CHMP2BIntron5 expression leads to gypsy activation
Having identified a genetic interaction between piRNA regulatory machinery and CHMP2BIntron5 expression in the fly eye, the molecular pathways involved in RV upregulation in the Drosophila FTD–ALS model expressing CHMP2BIntron5 were investigated. Levels of several RV genomic elements were examined by quantitative polymerase chain reaction (qPCR) in heads of flies expressing GMR > CHMP2BIntron5. Only insertions of the endogenous Drosophila RVs gypsy, gypsy-6 and tirant were observed to be significantly upregulated in the genome of GMR > CHMP2BIntron5 fly heads, with gypsy being the most strongly increased in abundance compared with wild-type and ago2 mutants, AGO2454/321 (Supplementary Material, Fig. S1A), a positive control known to regulate RVs (35,36). Individual RV elements can degenerate and lose ORFs from the RV element, but still be mobilized in a non-autonomous manner by intact RVs. To examine the abundance of degenerate gypsy elements in the presence of CHMP2BIntron5, we quantified the gypsy-genome components GAG, POL, ENV and LTR copy levels in the CHMP2BIntron5 expressing genome (Supplementary Material, Fig. S1B). A significant increase was observed only for the ENV (Fig. 1A and Supplementary Material, Fig. S1B) and LTR (Supplementary Material, Fig. S1B) sequences in the CHMP2BIntron5 expression background compared with the wild-type indicating a mobilization of a degenerate gypsy element containing ENV.
Given the increase of gypsy-ENV sequence in CHMP2BIntron5 flies (Fig. 2A), ENV protein levels in CHMP2BIntron5 expressing tissue were then examined. Immunoblotting using an anti-gypsy-ENV antibody (37) confirmed ENV to be present in CHMP2BIntron5 expressing and AGO2454/321 mutant heads in comparison to the wild-type heads (Fig. 2B), indicating a significant expression of the gypsy-ENV protein in the presence of CHMP2BIntron5.
Figure 2.
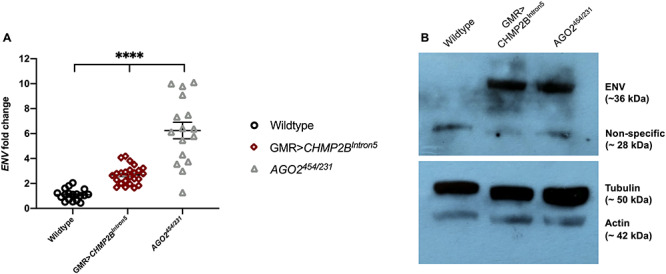
Increased gypsy-ENV expression is detected in pan-neuronally expressing CHMP2BIntron5 Drosophila. (A) Fold change in expression of gypsy-ENV measured in wild-type (+), GMR-Gal4:UAS-CHMP2BIntron5 (GMR > CHMP2BIntron5) and AGO2454/231 mutant fly heads by qPCR. Transcript levels are normalized to Rpl32 and fold-change values are relative to wild-type flies. Error bars indicate standard error (Kruskal–Wallis test, ****P < 0.0001). (B) Immunoblot showing ENV levels in wild-type, GMR > CHMP2BIntron5 and AGO2454/321 adult heads.
Gypsy mobilization is observed in vivo in CHMP2BIntron5 expressing neurons
To test whether gypsy is an active RV transposing physically in the presence of CHMP2BIntron5, we employed a reporter system based on RV integration into a genomic RV landing hotspot, known as ovo (38). We used a reporter system termed gypsy-TRAP to detect de novo insertions of gypsy [Fig. 3A; (28)]. In this system, a promoter-Gal4 construct is used to drive GFP in a tissue of interest, here OK6-Gal4, which expresses in motor neurons. In the same fly, Gal80 is expressed under the control of an α-tubulin-promoter repressing Gal4 in all tissues. Between the α-tubulin-promoter and the Gal80 element there is a section of the ovo promoter, a genomic hotspot for gypsy insertion events (37). If gypsy elements jump into the intervening hotspot, Gal80 expression is reduced and Gal4 is de-repressed, driving a GFP signal in motor neurons (Fig. 3A and B). To allow both CHMP2BIntron5 and CHMP2BWildtype expression in combination with the gypsy-TRAP system, a Gal4 independent expression system was employed. To achieve this, the LexA/LexAop system (39) was used to express CHMP2BWildtype and CHMP2BIntron5. To ensure genetic consistency for the initial number of genomic gypsy insertions prior to CHMP2B induced activation, both constructs were genomically site landed to ensure identical genomic backgrounds and equivalent expression levels of both CHMP2BWildtype and CHMP2BIntron5 proteins. Transposition was assayed in motor neurons of pan-neuronal expression of CHMP2BWildtype or CHMP2BIntron5 via nSyb-LexA (Fig. 3B). In the presence of Gal80 and absence of CHMP2BIntron5 expression, GFP is silenced and few motor neurons are labeled. When CHMP2BIntron5 is neuronally expressed, we observed GFP expression in multiple single motor neurons (Fig. 3B and C). In the presence of CHMP2BIntron5, the gypsy-TRAP highlights a significant increase in gypsy mobilization in CHMP2BIntron5 expressing cells compared with CHMP2BWildtype (Fig. 3B and C). Together with the findings described above, these results demonstrate that gypsy activity is higher in CHMP2BIntron5 expressing Drosophila neurons.
Figure 3.
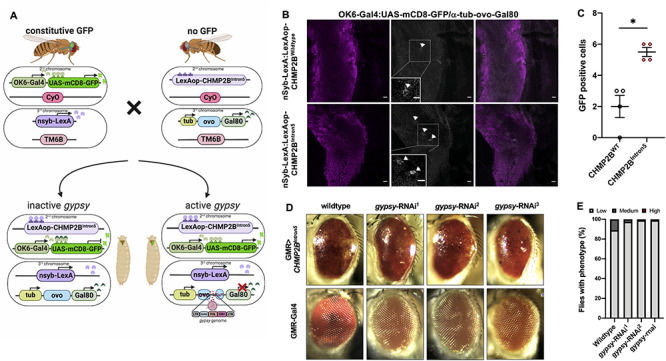
The gypsy-TRAP reporter confirms increased de novo insertion of gypsy in CHMP2BIntron5 expressing Drosophila brains. (A) Schematic of the gypsy-TRAP mobilization assay. De-repressed gypsy jumps into the hotspot ovo binding site and disrupts Gal80, triggering GFP expression in motor neurons. Image created with BioRender. (B) Pan neuronally driven (nSyb-LexA) CHMP2BIntron5 brains show increased GFP-positive cells compared with pan neuronally driven CHMP2BWildtype brains. GFP-positive cells represent gypsy mobilization events, which are limited to the motor neurons because of the OK6-Gal4 driver. Scale bar, 10 μm. (C) Quantification of GFP-labeled neurons observed in nSyb > CHMP2BWildtype and nSyb > CHMP2BIntron5 expressing Drosophila brains (t test, *P < 0.01). (D) Inhibition of gypsy via three different gypsy-RNAi (1, 2 and 3) rescued the degenerative eye phenotype caused by GMR-Gal4:UAS-CHMP2BIntron5 (GMR > CHMP2BIntron5). (E) Quantification of the eye phenotypes from (D) genotypes (n = 100).
Having identified an upregulation of gypsy-ENV and gypsy-mediated transposition events in CHMP2BIntron5 expressing neurons, we then asked whether it was possible to rescue the melanized eye phenotype caused by GMR > CHMP2BIntron5 via gypsy downregulation (Fig. 3D and E). Three different RNAi constructs targeting gypsy were tested for their ability to reverse melanization in fly eyes expressing CHMP2BIntron5 (40). Reducing gypsy via the three different RNAi constructs alleviated the melanization phenotype in the fly eye caused by CHMP2BIntron5 expression (Fig. 3D and E).
Inhibition of reverse transcriptase rescues neuronal aberrations caused by mammalian CHMP2BIntron5 expression
Having observed an amelioration of the melanized eye phenotype in Drosophila when gypsy was inhibited, we asked whether similar effects could be recapitulated in mammalian models. Neurons transfected with CHMP2BIntron5 develop a significant retraction of the dendritic arbor when compared with CHMP2BWildtype expressing controls [Fig. 4; (18)]. Stavudine (D4T) and lamivudine (3TC) are nucleoside analogue reverse transcriptase inhibitors that are widely used to inhibit human immunodeficiency virus replication in patients and are well tolerated as an anti-retroviral therapy (41). Primary neurons transfected with CHMP2BIntron5 showed a significant dendritic collapse phenotype compared with neurons transfected with CHMP2BWildtype. CHMP2BIntron5 expressing neurons develop a significant reduction in the length of the longest neuronal process (Fig. 4B, 40% reduction) and a reduction in total arbor size (Fig. 4C, 60% reduction) when compared with CHMP2BWIldtype expressing controls. This reduced complexity of the dendritic arbor was further analyzed using Sholl analysis, with a significant decrease in the cumulative number of intersections observed compared with wild-type controls (Fig. 4D). Administration of 10 μm of D4T (Stavudine) (48 h; Fig. 4) was sufficient to rescue these altered dendritic phenotypes. This observation was further confirmed with the administration of 10 μm 3TC (Lamivudine) (Supplementary Material, Fig. S2).
Figure 4.
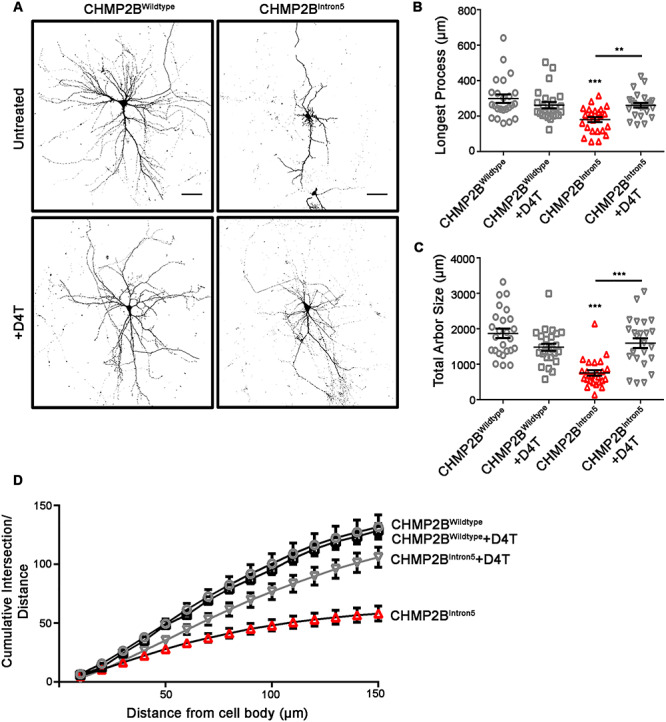
The retroviral reverse transcriptase inhibitor Stavudine (D4T) rescues dendritic collapse in mammalian neurons expressing CHMP2BIntron5. (A) Representative micrographs of mature neurons expressing FLAG-tagged CHMP2BWildtype or CHMP2BIntron5 in the absence and presence of D4T (Stavudine). Scale bar, 50 μm. Longest process length (B) and total arbor size (C) in CHMP2BIntron5 expressing neurons are significantly increased upon addition of D4T. (D) CHMP2BIntron5 presents a reduced cumulative branch number partially rescued by D4T. (B–D) One-way ANOVA with Tukey’s post-hoc comparing CHMP2BIntron5 and without D4T (**P < 0.01, ***P < 0.001).
We observe that manipulation of cellular RV silencing machinery rescues innate immune related melanization phenotypes in the Drosophila eye caused by CHMP2BIntron5 expression. Additionally inhibition of retroviral reverse transcriptase in mammalian neurons expressing CHMP2BIntron5 rescues dendritic collapse. Inhibition of gypsy via RNAi and administration of retroviral reverse transcriptase inhibitors D4T or 3TC was able to alleviate neurodegeneration-related phenotypes compared with controls (Figs 3D and E, Fig. 4; Supplementary Material, Fig. S2). Together, these data suggest that RV activity is increased in our Drosophila model and confirm that suppression of RV activation in both Drosophila and mammalian neurons reduces neurotoxicity, indicating RV suppression would be an attractive target for FTD–ALS treatment.
Discussion
Our observations using Drosophila as a model for the FTD-causing mutation CHMP2BIntron5 support an activation of the endogenous RV gypsy in CHMP2BIntron5 expressing Drosophila brains. Here, the RNA-silencing machinery Yb components, including BoYb, SoYb, Vret and piwi, were identified as dominant modifiers of CHMP2BIntron5 toxicity. Recent reports have found levels of RV encoded reverse transcriptase activity in serum and cerebrospinal fluid of FTD–ALS patients (30,42–45). Other studies have found in postmortem ALS tissue the presence of ENV, encoded by hERV-K, a human RV akin to Drosophila-gypsy (26,46). These findings have been replicated in a TDP-43 expressing Drosophila model of FTD–ALS (28,47). In this investigation, gypsy, gypsy-6 and tirant insertions were increased in the presence of CHMP2BIntron5 among other RVs suggesting differential regulation of RVs. We focused our study on the well characterized RV gypsy. Here, a significant increase in gypsy-ENV transcript, as well as an upregulation of the glycoprotein ENV, is found in CHMP2BIntron5 expressing Drosophila heads. An increase in RV transcripts identified through a transcriptome analysis from C9orf72-affected patients has been previously observed (48). In an in vitro study to confirm hERV-K-ENV pathogenicity, the hERV-K-ENV ORF was transfected into human neuronal cells, which induced a decrease in cell number (26). In the same study, a transgenic mouse expressing hERV-K-ENV protein under a neuronal promoter was constructed and these animals developed an evident motor neuron degeneration, DNA damage and cortical thinning of the rostral part of the motor cortex (26). This suggests that the activation of these elements may be conserved and present among different FTD–ALS-causative mutations, though not all patients show elevated levels of hERV-K. RV elements have been characterized as TDP-43 targets at the DNA level (47) and an increase in RV expression found in ALS patients correlates with TDP-43 pathology (49). Although CHMP2BIntron5 patients do not have apparent TDP-43 inclusions in the cytoplasm (12), manipulation of functionally associated ESCRT proteins can cause TDP-43 aggregation (50). We therefore cannot rule out a role for compromised TDP-43 function in CHMP2BIntron5 associated RV re-activation.
The suppression of RVs through reverse transcriptase inhibitors can ameliorate neurological defects associated with aging and FTD–ALS in Drosophila models (47,51). Here, we provide evidence that gypsy knockdown alleviates neuronal defects in our Drosophila model of FTD caused by the CHMP2BIntron5 mutation. In our rat primary neuron model of FTD caused by CHMP2BIntron5 expression, reverse transcriptase inhibition is proposed to block mRNA production from RV elements via the inhibition of POL activity and the subsequent production of further GAG, POL and ENV proteins. Using two clinically well-characterized antiviral reverse transcriptase inhibitors, stavudine (D4T) and lamivudine (3TC), we were able to prevent the development of dendritic collapse in rat primary neurons expressing CHMP2BIntron5 (Fig. 4; Supplementary Material, Fig. S2). Preventing RV mobilization may help to limit neuronal and/or glial cell death in FTD–ALS. Recent studies have pointed to a role for TDP-43 in repressing RVs and retrotransposons (RTs), which also act via reverse transcriptase POL activity, both in human and Drosophila DNA (47,51–53). Around 50% of FTD patients do not have TDP-43 misregulation while 97% of ALS patients have TDP-43 aggregation (54,55), suggesting that only TDP-43-positive patients would benefit from RV inhibition. CHMP2BIntron5-affected patients are TDP-43-negative (7). In our Drosophila model expressing CHMP2BIntron5, RV disruption may be caused by other factors and not only by TDP-43 disturbance. Several RNA binding proteins (RBPs) linked to ALS, including MATR3, TDP-43, TIA1, hnRNPA1 and TAF15, are found to bind and regulate the RT LINE1 RNA elements (56). This suggests that disruption of RBP function might dysregulate RV/RT repression. Thus, CHMP2BIntron5-affected patients could also benefit from RV inhibition, as our data suggests. Our work provides functional evidence for a conserved mechanism between Drosophila and mammalian models of FTD–ALS where RV inhibition provides an amelioration of the neurodegenerative condition via mechanisms that are yet to be discovered.
Materials and Methods
Fly stocks and maintenance
Flies were raised at 25°C on standard yeast, sugar and agar medium on a 12 h light:dark cycle. Unless otherwise stated, Canton-S was used for control. The following Drosophila lines were obtained from the Bloomington Stock Center, Indiana: UAS-BoYb (BL#28446), UAS-BoYb-RNAi (BL#62432), piwi1 (BL#43637), piwi2 (BL#43319), UAS-piwi-RNAi (BL#33724), UAS-SoYb (BL#26961), UAS-SoYb-RNAi (BL#36881), UAS-Vret (BL#22204), UAS-Vret-RNAi (BL#38212), AGO2454 (BL#36522), AGO2321 (BL#36511), glass multiple reporter GMR-Gal4 (BL#1104), neuronal synaptobrevin (nSyb)-LexA (BL#52817), OK6-Gal4 (Cahir O’Kane, University of Cambridge, UK), UAS-mCD8-GFP (BL#32186). For the genetic screen, lines containing GMR-Gal4 and UAS-CHMP2BIntron5 were recombined into the second chromosome as described elsewhere (15). gypsy-TRAP transgenic flies were a gift from Josh Dubnau (28). gypsy-RNAi transgenic flies were obtained from Jeng Pin (40). For flies used in gDNA extraction experiments, UAS-CHMP2BIntron5 and UAS-CHMP2BWildtype (15) were maintained in the same w1118 background and crossed to GMR-GAL4 to ensure identical genetic backgrounds.
Genetic interaction studies
To quantify the CHMP2BIntron5 eye phenotype, the classification was divided into low, medium or high levels of melanization. GMR-GAL4::UAS-CHMP2BIntron5 (GMR > CHMP2BIntron5) virgin females were crossed to mutant and transgenic stocks. Eyes were imaged using a camera (AxioCam Erc 5 s; Carl Zeiss, Germany) on a dissecting scope (Stemi 2000-C; Carl Zeiss).
Generation of transgenic fly lines
To generate LexAop-CHMP2BWildtype and LexAop-CHMP2BIntron5 transgenic flies, the primers listed in Supplementary Material, Table S1 were used to amplify CHMP2BIntron5 and CHMP2BWildtype and cloned into a pJFRC19-13XLexAop2-IVS-myr::GFP vector (57) (Addgene, USA, Cat No. 26224) via the BglII and Xba1 cloning sites. Constructs were then sequenced and inserted into the Drosophila genome at the attP40 insertion site via C31-mediated site-specific attP integration. Integration at the attP40 site on the second chromosome was achieved by microinjection into the stock: y,w,M(eGFP, vas-int, dmRFP)ZH-2A; P{CaryP}attP40. Microinjections were carried out by the Cambridge Microinjection Service (UK).
Immunohistochemistry
Drosophila immunohistochemistry was performed as described elsewhere (17). Primary antibodies used were: mouse anti-elav (DSHB 9F8A9, 1:50) and FluoTag®-X4 anti-GFP (NanoTag Biotechnologies, Germany, 1:1000). Primary antibodies were incubated with preparations overnight at 4°C. The following day, after washing with PBT they were incubated with the appropriate secondary antibodies for 1 h at room temperature before washing with PBT and mounting with Vectashield Mounting Medium (Vector Labs, USA). Control larvae were stained in the same solutions as experimental larvae. For rat primary neuron immunohistochemistry, cells were fixed and stained as previously described (58) using anti-FLAG (Sigma, USA, M2 clone, 1:1000). Primary antibodies were incubated overnight at 4°C. Corresponding Alexafluor secondary antibodies (Thermo Scientific, USA, 1:500) were incubated for 1 h at room temperature before mounting with Fluoromount (Sigma).
Confocal microscopy and image analysis
For imaging Drosophila tissue, confocal images were acquired using a ZEISS LSM 880 confocal microscope using a 20× or 40× objectives using Zeiss filter sets for Alexa 488/561/633. Z-stack images of larvae brains were obtained using 20× or 40× NA oil objective. Images stacks were merged and processed using Fiji. For imaging rat primary neurons, images were collected on an inverted Zeiss microscope (880) with 20× Plan Neofluar objectives using Zeiss filter sets for DAPI and Alexa 488/546. Images were taken at an aspect ratio of 1024 × 1024. Images of neurons were traced using the NeuronJ plugin in ImageJ (1.6.0). Individual traces were saved, thresholded and Sholl analysis was conducted using the Sholl plugin.
qPCR data analysis
gDNA was extracted using Puregene Core Kit A (Qiagen). SYBR® green assays were used to perform the qPCR analysis on a QuantStudio3 Real Time PCR System (Thermo Fisher Scientific, USA). Samples were run in triplicate and normalized to rpl32 expression. Relative expression of genes was determined by the 2−∆∆Ct method (59). Primers are listed in Supplementary Material, Table S1.
Protein extraction and immunoblot
Fifty adult fly heads were homogenized in 30 μL of 2× Laemmli loading buffer. After boiling, samples were run on a 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Biorad, USA) and transferred to PVDF membrane (Invitrogen, USA). Primary antibodies used include: mouse anti-ENV (37), anti-ß-Actin (Proteintech, USA, 60008–1-Ig, 7D2C10, 1:8000) and anti-ß-tubulin E7 (DSHB, 1:5000).
Culture of primary neurons
Timed-mated female Wistar rats (Charles River, UK) (RRID:RGD_737929) were maintained in accordance with the UK Animals (Scientific Procedures) Act (1986). Cortices were dissected from postnatal day 1 (P1) mixed sex rat pups. Animals were euthanized using pentobarbital injection followed by cervical dislocation, according to Home Office guidelines. Cortical cell suspensions were obtained as previously (60) described and cytosine arabinoside (AraC, 2.4 μm final concentration) was added to the growth medium at 1 days in vitro (DIV). Neurons were transfected at 12 DIV with Lipofectamine 2000 (11668019, Thermo Scientific) with either FLAG-tagged CHMP2BWildtype or CHMP2BIntron5, described previously (61) and treated with either or stavudine or lamivudine for 48 h (10 μm).
Statistical analysis
The statistical data were performed using GraphPad Prism (6.01). Data are presented as mean values, from at least three independent biological replicates, with error bars representing the standard error of mean. All details of the tests used were outlined within each figure legend.
Supplementary Material
Acknowledgements
We thank Josh Dubnau (Cold Spring Harbor, NY, USA) for the kind gift of gypsy-TRAP and Peng Jin (Atlanta, GA, USA) for the kind gift of gypsy-RNAi stocks. We thank Joseph G. Gall (Baltimore, MD, USA) for providing the monoclonal ENV antibody. We thank Stavroula Petridi (Warwick, UK) for providing the LexAop plasmid. We thank the Bloomington Drosophila Stock Center (Indiana, USA) for providing Drosophila stocks. We thank the Bioscience Technology Facility at the University of York for providing access to confocal microscopes.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Motor Neurone Disease Association, grant reference Sweeney/Oct15/884-792 awarded to S.T.S. and was partly funded by The Wellcome Trust [grant number: 204829] through the Centre for Future Health (CFH) awarded to C.U. at the University of York.
Author Contributions
L.F.A. and S.T.S designed research; L.F.A and C.U. performed research; L.F.A and C.U. analysed data; and L.F.A and S.T.S wrote the paper.
References
- 1. Ferrari R., Forabosco P., Vandrovcova J., Botia J.A., Guelfi S., Warren J.D., Consortium U.K.B.E., Momeni P., Weale M.E., Ryten M. et al. (2016) Frontotemporal dementia: insights into the biological underpinnings of disease through gene co-expression network analysis. Mol. Neurodegener., 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim E.J., Sidhu M., Gaus S.E., Huang E.J., Hof P.R., Miller B.L., DeArmond S.J. and Seeley W.W. (2012) Selective frontoinsular von economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb. Cortex, 22, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burrell J.R., Halliday G.M., Kril J.J., Ittner L.M., Gotz J., Kiernan M.C. and Hodges J.R. (2016) The frontotemporal dementia-motor neuron disease continuum. Lancet, 388, 919–931. [DOI] [PubMed] [Google Scholar]
- 4. Ferrari R., Kapogiannis D., Huey E.D. and Momeni P. (2011) FTD and ALS: a tale of two diseases. Curr. Alzheimer Res., 8, 273–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ling S.C., Polymenidou M. and Cleveland D.W. (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron, 79, 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zucchi E., Ticozzi N. and Mandrioli J. (2019) Psychiatric symptoms in amyotrophic lateral sclerosis: beyond a motor neuron disorder. Front. Neurosci., 13, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T. et al. (2005) Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet., 37, 806–808. [DOI] [PubMed] [Google Scholar]
- 8. Zee J., Urwin H., Engelborghs S., Bruyland M., Vandenberghe R., Dermaut B., De Pooter T., Peeters K., Santens P., De Deyn P.P. et al. (2008) CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet., 17, 313–322. [DOI] [PubMed] [Google Scholar]
- 9. Cox L.E., Ferraiuolo L., Goodall E.F., Heath P.R., Higginbottom A., Mortiboys H., Hollinger H.C., Hartley J.A., Brockington A., Burness C.E. et al. (2010) Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS). PLoS One, 5, e9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narain P., Pandey A., Gupta S., Gomes J., Bhatia R. and Vivekanandan P. (2018) Targeted next-generation sequencing reveals novel and rare variants in Indian patients with amyotrophic lateral sclerosis. Neurobiol. Aging, 71(265), e269–265 e214. [DOI] [PubMed] [Google Scholar]
- 11. Blitterswijk M., Vlam L., Es M.A., Pol W.L., Hennekam E.A., Dooijes D., Schelhaas H.J., Kooi A.J., Visser M., Veldink J.H. et al. (2012) Genetic overlap between apparently sporadic motor neuron diseases. PLoS One, 7, e48983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parkinson N., Ince P.G., Smith M.O., Highley R., Skibinski G., Andersen P.M., Morrison K.E., Pall H.S., Hardiman O., Collinge J. et al. (2006) ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology, 67, 1074–1077. [DOI] [PubMed] [Google Scholar]
- 13. Babst M., Katzmann D.J., Snyder W.B., Wendland B. and Emr S.D. (2002) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell, 3, 283–289. [DOI] [PubMed] [Google Scholar]
- 14. Urwin H., Authier A., Nielsen J.E., Metcalf D., Powell C., Froud K., Malcolm D.S., Holm I., Johannsen P., Brown J. et al. (2010) Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet., 19, 2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmad S.T., Sweeney S.T., Lee J.A., Sweeney N.T. and Gao F.B. (2009) Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc. Natl. Acad. Sci. U S A., 106, 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu Y., Zhang Z., Sun D., Sweeney S.T. and Gao F.B. (2013) Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell, 52, 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. West R.J., Lu Y., Marie B., Gao F.B. and Sweeney S.T. (2015) Rab8, POSH, and TAK1 regulate synaptic growth in a drosophila model of frontotemporal dementia. J. Cell Biol., 208, 931–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. West R.J.H., Ugbode C., Gao F.B. and Sweeney S.T. (2018) The pro-apoptotic JNK scaffold POSH/SH3RF1 mediates CHMP2BIntron5-associated toxicity in animal models of frontotemporal dementia. Hum. Mol. Genet., 27, 1382–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H. and Siomi M.C. (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the drosophila genome. Genes Dev., 20, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Volff J.N. (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays, 28, 913–922. [DOI] [PubMed] [Google Scholar]
- 21. Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R. and Hannon G.J. (2009) Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. Cell, 137, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Senti K.A. and Brennecke J. (2010) The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet., 26, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Golden D.E., Gerbasi V.R. and Sontheimer E.J. (2008) An inside job for siRNAs. Mol. Cell, 31, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Douville R.N. and Nath A. (2017) Human endogenous retrovirus-K and TDP-43 expression bridges ALS and HIV neuropathology. Front. Microbiol., 8, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garson J.A., Usher L., Al-Chalabi A., Huggett J., Day E.F. and McCormick A.L. (2019) Quantitative analysis of human endogenous retrovirus-K transcripts in postmortem premotor cortex fails to confirm elevated expression of HERV-K RNA in amyotrophic lateral sclerosis. Acta Neuropathol. Commun., 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li W., Lee M.H., Henderson L., Tyagi R., Bachani M., Steiner J., Campanac E., Hoffman D.A., Geldern G., Johnson K. et al. (2015) Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med., 7, 307ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang Y.H., Keegan R.M., Prazak L. and Dubnau J. (2019) Cellular labeling of endogenous retrovirus replication (CLEVR) reveals de novo insertions of the gypsy retrotransposable element in cell culture and in both neurons and glial cells of aging fruit flies. PLoS Biol., 17, e3000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., Prazak L., Chatterjee N., Gruninger S., Krug L., Theodorou D. and Dubnau J. (2013) Activation of transposable elements during aging and neuronal decline in drosophila. Nat. Neurosci., 16, 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morera A.A., Ahmed N.S. and Schwartz J.C. (2019) TDP-43 regulates transcription at protein-coding genes and Alu retrotransposons. Biochim. Biophys. Acta Gene Regul. Mech., 1862, 194434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andrews W.D., Tuke P.W., Al-Chalabi A., Gaudin P., Ijaz S., Parton M.J. and Garson J.A. (2000) Detection of reverse transcriptase activity in the serum of patients with motor neurone disease. J. Med. Virol., 61, 527–532. [DOI] [PubMed] [Google Scholar]
- 31. Gold J., Rowe D.B., Kiernan M.C., Vucic S., Mathers S., Eijk R.P.A., Nath A., Garcia Montojo M., Norato G., Santamaria U.A. et al. (2019) Safety and tolerability of Triumeq in amyotrophic lateral sclerosis: the lighthouse trial. Amyotroph. Lateral Scler. Frontotemporal Degener., 20, 595–604. [DOI] [PubMed] [Google Scholar]
- 32. Handler D., Olivieri D., Novatchkova M., Gruber F.S., Meixner K., Mechtler K., Stark A., Sachidanandam R. and Brennecke J. (2011) A systematic analysis of drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J., 30, 3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Handler D., Meixner K., Pizka M., Lauss K., Schmied C., Gruber F.S. and Brennecke J. (2013) The genetic makeup of the drosophila piRNA pathway. Mol. Cell, 50, 762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toth K.F., Pezic D., Stuwe E. and Webster A. (2016) The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol., 886, 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R. et al. (2008) An endogenous small interfering RNA pathway in drosophila. Nature, 453, 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z. et al. (2008) Endogenous siRNAs derived from transposons and mRNAs in drosophila somatic cells. Science, 320, 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song S.U., Gerasimova T., Kurkulos M., Boeke J.D. and Corces V.G. (1994) An env-like protein encoded by a drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev., 8, 2046–2057. [DOI] [PubMed] [Google Scholar]
- 38. Dej K.J., Gerasimova T., Corces V.G. and Boeke J.D. (1998) A hotspot for the drosophila gypsy retroelement in the ovo locus. Nucleic Acids Res., 26, 4019–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai S.L. and Lee T. (2006) Genetic mosaic with dual binary transcriptional systems in drosophila. Nat. Neurosci., 9, 703–709. [DOI] [PubMed] [Google Scholar]
- 40. Tan H., Qurashi A., Poidevin M., Nelson D.L., Li H. and Jin P. (2012) Retrotransposon activation contributes to fragile X premutation rCGG-mediated neurodegeneration. Hum. Mol. Genet., 21, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krogstad P., Lee S., Johnson G., Stanley K., McNamara J., Moye J., Jackson J.B., Aguayo R., Dieudonne A., Khoury M. et al. (2002) Nucleoside-analogue reverse-transcriptase inhibitors plus nevirapine, nelfinavir, or ritonavir for pretreated children infected with human immunodeficiency virus type 1. Clin. Infect. Dis., 34, 991–1001. [DOI] [PubMed] [Google Scholar]
- 42. MacGowan D.J., Scelsa S.N., Imperato T.E., Liu K.N., Baron P. and Polsky B. (2007) A controlled study of reverse transcriptase in serum and CSF of HIV-negative patients with ALS. Neurology, 68, 1944–1946. [DOI] [PubMed] [Google Scholar]
- 43. McCormick A.L., Brown R.H. Jr., Cudkowicz M.E., Al-Chalabi A. and Garson J.A. (2008) Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology, 70, 278–283. [DOI] [PubMed] [Google Scholar]
- 44. Steele A.J., Al-Chalabi A., Ferrante K., Cudkowicz M.E., Brown R.H. Jr. and Garson J.A. (2005) Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology, 64, 454–458. [DOI] [PubMed] [Google Scholar]
- 45. Viola M.V., Frazier M., White L., Brody J. and Spiegelman S. (1975) RNA-instructed DNA polymerase activity in a cytoplasmic particulate fraction in brains from Guamanian patients. J. Exp. Med., 142, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Douville R., Liu J., Rothstein J. and Nath A. (2011) Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol., 69, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krug L., Chatterjee N., Borges-Monroy R., Hearn S., Liao W.W., Morrill K., Prazak L., Rozhkov N., Theodorou D., Hammell M. et al. (2017) Retrotransposon activation contributes to neurodegeneration in a drosophila TDP-43 model of ALS. PLoS Genet., 13, e1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. et al. (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci., 18, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tam O.H., Rozhkov N.V., Shaw R., Kim D., Hubbard I., Fennessey S., Propp N., Consortium N.A., Fagegaltier D., Harris B.T. et al. (2019) Postmortem cortex samples identify distinct molecular subtypes of ALS: retrotransposon activation, oxidative stress, and activated glia. Cell Rep., 29, 1164–1177 e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerod L., Fisher E.M., Isaacs A., Brech A., Stenmark H. and Simonsen A. (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol., 179, 485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wood J.G., Jones B.C., Jiang N., Chang C., Hosier S., Wickremesinghe P., Garcia M., Hartnett D.A., Burhenn L., Neretti N. et al. (2016) Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in drosophila. Proc. Natl. Acad. Sci. U S A., 113, 11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu E.Y., Russ J., Cali C.P., Phan J.M., Amlie-Wolf A. and Lee E.B. (2019) Loss of nuclear TDP-43 is associated with decondensation of LINE retrotransposons. Cell Rep., 27, 1409–1421 e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Romano G., Klima R. and Feiguin F. (2020) TDP-43 prevents retrotransposon activation in the Drosophila motor system through regulation of Dicer-2 activity. BMC Biology, 18, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mandrioli J., Mediani L., Alberti S. and Carra S. (2020) ALS and FTD: where RNA metabolism meets protein quality control. Semin. Cell Dev. Biol., 99, 183–192. [DOI] [PubMed] [Google Scholar]
- 55. Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C. et al. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci., 14, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Attig J., Agostini F., Gooding C., Chakrabarti A.M., Singh A., Haberman N., Zagalak J.A., Emmett W., Smith C.W.J., Luscombe N.M. et al. (2018) Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell, 174, 1067–1081 e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pfeiffer B.D., Ngo T.B., Hibbard K.L., Murphy C., Jenett A., Truman J.W. and Rubin G.M. (2010) Refinement of tools for targeted gene expression in drosophila. Genetics, 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ugbode C.I., Smith I., Whalley B.J., Hirst W.D. and Rattray M. (2017) Sonic hedgehog signalling mediates astrocyte crosstalk with neurons to confer neuroprotection. J. Neurochem., 142, 429–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Livak K.J. and Schmittgen T.D. (2001) Analysis of relative expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 60. Suman R., Smith G., Hazel K.E., Kasprowicz R., Coles M., O'Toole P. and Chawla S. (2016) Label-free imaging to study phenotypic behavioural traits of cells in complex co-cultures. Sci. Rep., 6, 22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee J.A., Beigneux A., Ahmad S.T., Young S.G. and Gao F.B. (2007) ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol., 17, 1561–1567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


