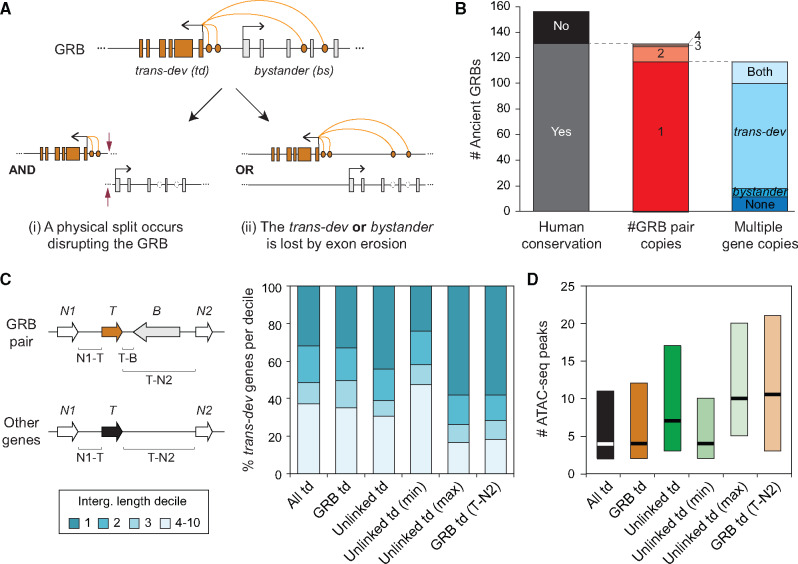
Fig. 1.
Fates of ancient GRBs after the two WGDs in vertebrates. (A) Genetic redundancy allows the dismantling of GRBs after WGD. Two scenarios are depicted: (i) The GRB microsyntenic association is disrupted by a break point, in which both the trans-dev and the bystander genes may be maintained, but in different genomic locations; This scenario would impact the regulation of the trans-dev gene. (ii) The GRB microsyntenic association is dismantled by the differential loss of either the trans-dev or the bystander gene. The loss can occur by a large deletion of the genomic locus or by pseudogenization and “exon erosion.” Although the former would impact the regulation of the trans-dev when the bystander is lost, the latter would not. (B) Summary of the fates of the 156 studied ancient GRB pairs present by the last common ancestor of chordates. Human conservation, whether the human genome has conserved at least one linked copy or not of the ancient GRB associations; #GRB pair copies, for those conserved, the number of copies of GRB pairs maintained in human (1–4); Multiple gene copies, for those GRB pairs in single copy, in how many cases, there are multiple ohnologs for both the trans-dev and bystander genes (i.e., not linked), only for the trans-dev or bystander gene, or for none. (C) Percent of trans-dev (T) genes of different types that have at least one intergenic region (N1-T and/or T-B [for ancient GRB pairs] or T-N2 [for other trans-dev genes]) within the first, second, third, or another decile of intergenic region lengths genome-wide (i.e., trans-dev genes in decile 1 have at least one intergenic region whose length is among the top 10% of all intergenic regions). All td, all trans-dev genes linked to at least one non-trans-dev gene (n = 745); GRB td, trans-dev genes that are part of conserved ancient GRB pairs (n = 103); Unlinked td, trans-dev ohnologs from ancient GRBs that are not linked to the ancient bystander gene (n = 171); Unlinked td (max/min), the unlinked trans-dev ohnolog with the largest/smallest intergenic region (n = 107); GRB td (T-N2), for these cases, the distance from the trans-dev gene in a conserved ancient GRB pair to the gene after the bystander (gene N2) is considered as the only intergenic distance (n = 103). Genes are only counted once in each category and cases for which the two downstream neighbors (N1 and N2) are not present are not considered. All lengths for each category are provided in supplementary table S3, Supplementary Material online. The use of N1 and N2 to label gene neighbors does not imply that these genes are the ohnologs of the neighboring genes flanking a trans-dev gene before WGDs. (D) For each type of trans-dev gene, number of ATAC-seq peaks found in the intergenic region with the highest number of peaks, except for “GRB td (T-N2),” where only the number of peaks between the trans-dev and the gene N2 is considered (as in C; n = 655, 82, 131, 78, 78, and 82, respectively).