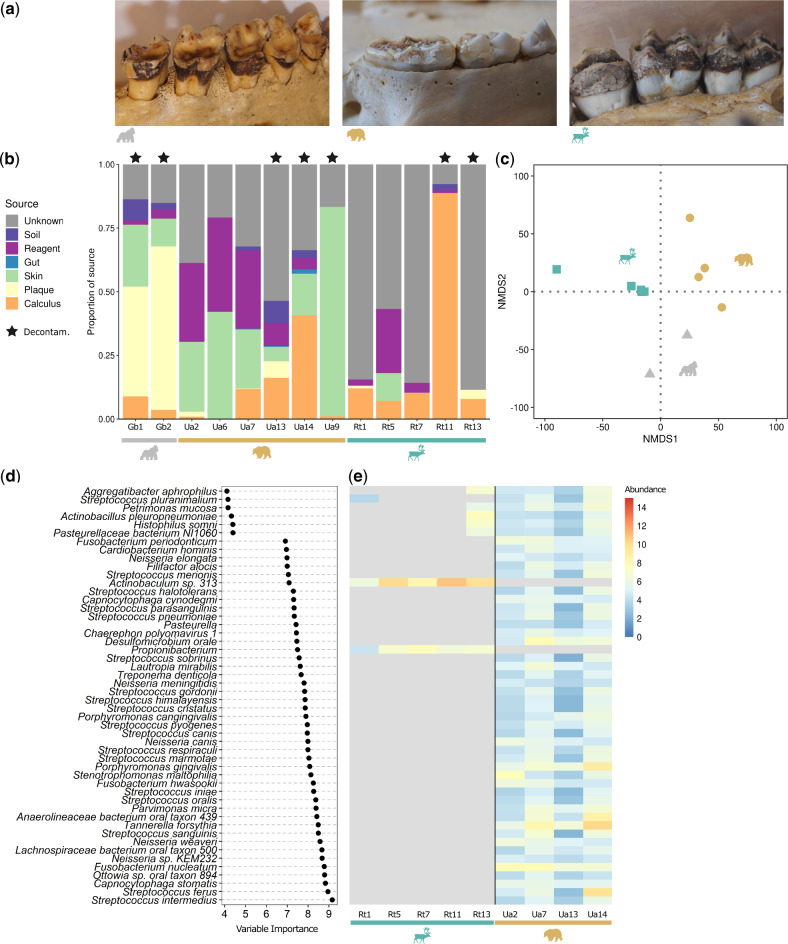
Fig. 1.
Dental calculus of nonhuman mammals shows an oral microbiome signature and contains host-specific taxa. (a) Dental calculus on the healthy teeth of a gorilla (Gorilla beringei), brown bear (Ursus arctos), and reindeer (Rangifer tarandus) specimen (left-to-right, respectively). Note these are representative photos of specimens in the museum collections. (b) Proportions of source contributions to the microbial communities (identified taxonomically at the species and genus level) of the dental calculus samples estimated by SourceTracker. Stars above bars indicate samples in which surface decontamination (UV and/or EDTA wash) was performed before DNA extraction. (c) NMDS ordination on Euclidean distance matrix of CLR normalized microbial abundances of taxa in samples from healthy teeth, colored by host species. NMDS stress: 0.107. (d) Random forest variable importance plot of the 30 most discriminatory taxa comparing bear and reindeer samples from healthy teeth, based on presence/absence of microbial taxa after contamination filtering. (e) CLR normalized abundance of the top 30 taxa in (d) in the bear and reindeer samples from healthy teeth. Taxa that were not detected in a sample are colored gray. Sample Ua9 from a carious bear was excluded in the analyses for (c–e), see supplementary figure S7, Supplementary Material online, for analysis with Ua9.