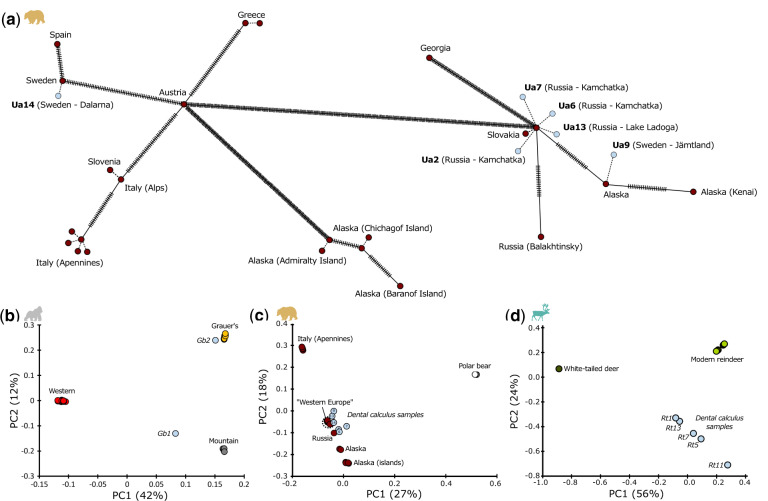
Fig. 5.
Host population genetic structure can be reconstructed from dental calculus. (a) mtDNA haplotype network for brown bears. Each circle represents a sample, with ticks on the connecting lines showing the number of base pair substitutions between the haplotypes. Dotted lines represent identical haplotypes or in the case of dental calculus samples (shown in light blue), the predicted most closely related haplotype. Sample labels include country and locality of specimen collection. Note that dental calculus samples of Swedish bears cluster with both eastern (Russia) and western (Spain) modern genomes, consistent with the history of postglacial colonization of Sweden. (b–d) PCA of modern high-coverage genomes and low-coverage dental calculus samples projected onto the modern genomes for gorilla (b), brown bear (c), and reindeer (d). Samples cluster together with their respective species. Dental calculus gorilla samples cluster most closely to their subspecies of origin. Dental calculus brown bear samples from western Europe and Russia cluster with the modern genomes from the “western Europe” clade (consisting of Spain, Greece, Slovenia, Italy [Alps], Sweden, and Slovakia) and the Russian clade, respectively. Dental calculus reindeer samples are clearly separated from the white-tailed deer outgroup.