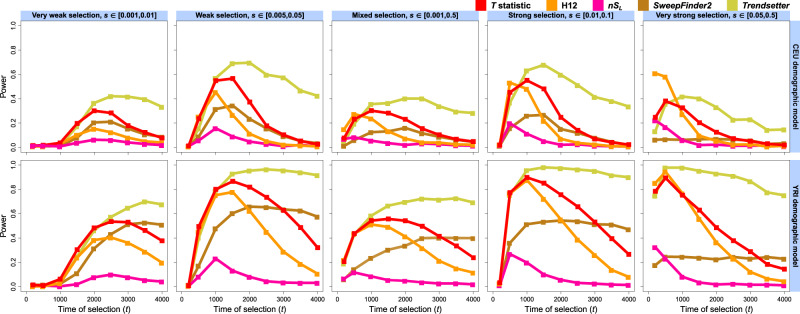
Fig. 3.
Powers of the T statistic and other sweep detection methods—H12, nSL, SweepFinder2, and Trendsetter—at the 1% FPR to detect soft selective sweeps from selection on standing variation on ν = 4 distinct sweeping haplotypes beginning at times generations prior to sampling, for the European CEU (top) and sub-Saharan African YRI (bottom) human demographic models inferred with smc++. Analysis data consisted of phased haplotypes of length 1 Mb, with 1,000 replicates for each distinct scenario. Selective sweeps were simulated for five ranges of selection coefficients (s) spanning very weak to very strong, with s for each replicate drawn uniformly at random (from a log scale for s drawn across orders of magnitude). All inferences used a spectrum of K = 10 for likelihood computations.