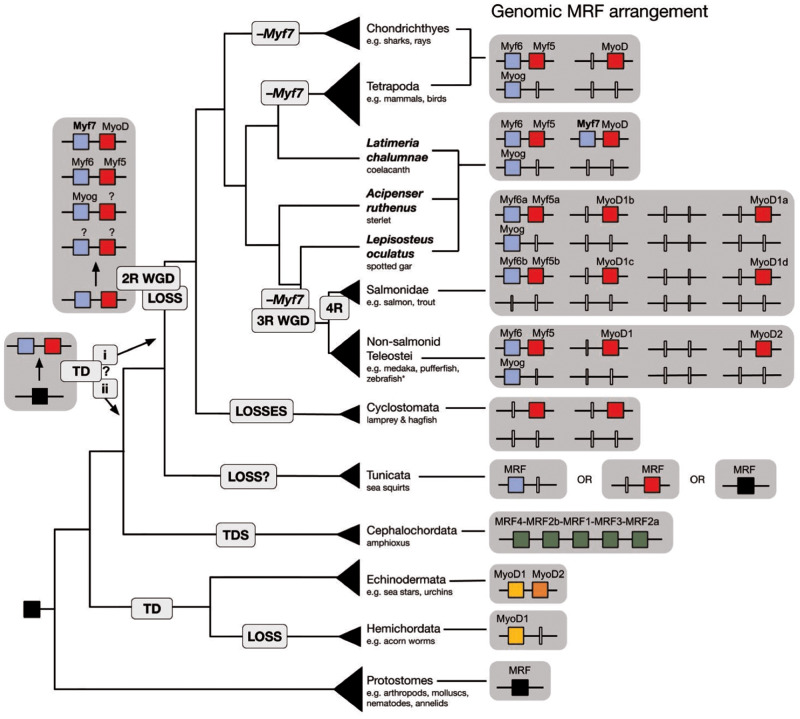
Fig. 2.
Schematic of bilaterian MRF evolution. Genomic MRF arrangement (right) for taxonomic groups (species cladogram, left) included in this study highlighting the vertebrate species that have retained Myf7 (coelacanth, sterlet, and spotted gar, bold text), as well as duplications and losses in several lineages. Retained MRFs are represented by colored boxes, whereas genes inferred to have been lost are represented by white vertical lines. Genes clustered in the genome are joined by black lines. The uncertainty as to the timing of the vertebrate early–late tandem duplication in the vertebrate (i) or Olfactores (ii) ancestor is denoted by the “?.” *Zebrafish has lost its MyoD2 paralog but is otherwise like the rest of nonsalmonid teleosts. TD, tandem duplication; 2R WGD, two rounds of whole-genome duplication; 3R WGD, teleost-specific third round of whole-genome duplication; 4R, salmonid-specific fourth round of whole-genome duplication; –Myf7, inferred loss of Myf7.