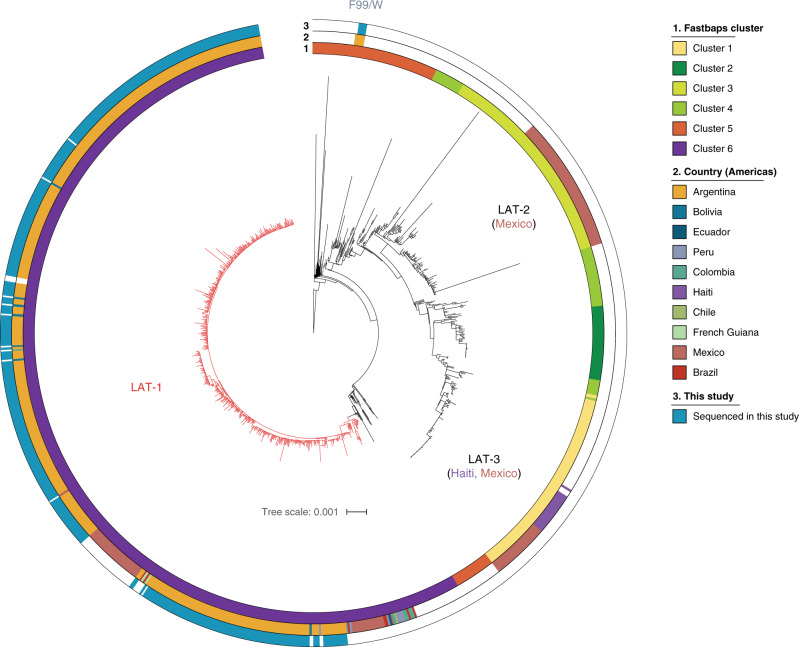
Fig. 2. The vast majority of Argentinian 7PET V. cholerae O1 are members of the LAT-1 sub-lineage.
A maximum-likelihood phylogeny produced for the 7PET lineage by calling SNVs between V. cholerae genome sequences and the N16961 reference genome sequence, rooted on M66, and excluding SNVs located in potentially recombined regions of the chromosome, for a total of 7,556 SNVs. Country of origin for genomes from the Americas are reported. All but four of the Argentinian 7PET genomes sequenced in this study are members of LAT-1 (in red). Clusters were determined using Fastbaps from an alignment of 3,874 parsimony-informative SNVs and used to delineate the LAT-1 sub-lineage (in red). Scale bar denotes the number of mutations per variable site.