Figure 1.
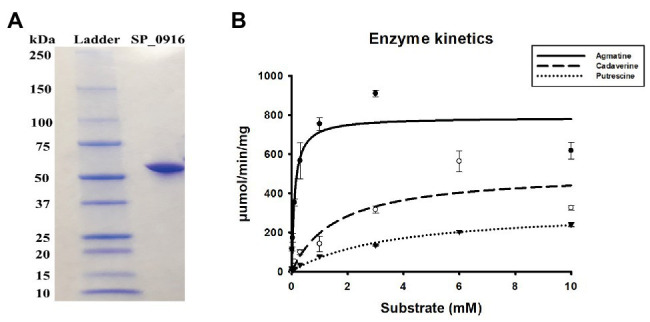
Gel electrophoresis and enzyme kinetics of SP_0916. (A) Overexpressed and purified recombinant SP_0916 was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie Brilliant Blue R-250 for visualization of the 54 KDa protein. The ladder lane on the left is the protein marker showing the molecular mass standards. (B) Enzyme kinetics for the conversion of arginine to agmatine, lysine to cadaverine, and ornithine to putrescine were performed in triplicate using liquid chromatography-mass spectrometry (LC-MS). The change in velocity with increase in substrate concentrations was fitted with Sigma Plot v.12 to estimate the kinetic parameters using the Michaelis-Menten equation by non-linear regression method. The K m of SP_0916 with the substrates is ornithine (3.55 ± 0.28 mM) > lysine (1.61 ± 0.28 mM) > arginine (0.11 ± 0.02 mM), indicating that arginine is the preferred substrate of this decarboxylase. High catalytic efficiency (kcat/Km) for the SP_0916-catalyzed conversion of arginine to agmatine (4.0 × 105 min−1mM−1) compared to low kcat/Km for conversion of lysine to cadaverine (1.7 × 104 min−1mM−1) and ornithine to putrescine (4.8 × 103 min−1mM−1) show that arginine is the preferred substrate for SP_0916. kcat/Km values are obtained using the means for each kinetic parameter.
